Blunt Snout Bream (Megalobrama amblycephala) MyD88 and TRAF6: characterisation, comparative homology modelling and expression
- PMID: 25830478
- PMCID: PMC4425005
- DOI: 10.3390/ijms16047077
Blunt Snout Bream (Megalobrama amblycephala) MyD88 and TRAF6: characterisation, comparative homology modelling and expression
Abstract
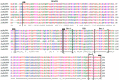
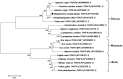
MyD88 and TRAF6 play an essential role in the innate immune response in most animals. This study reports the full-length MaMyD88 and MaTRAF6 genes identified from the blunt snout bream (Megalobrama amblycephala) transcriptome profile. MaMyD88 is 2501 base pairs (bp) long, encoding a putative protein of 284 amino acids (aa), including the N-terminal DEATH domain of 78 aa and the C-terminal TIR domain of 138 aa. MaTRAF6 is 5474 bp long, encoding a putative protein of 542 aa, including the N-terminal low-complexity region, RING domain (40 aa), a coiled-coil region (64 aa) and C-terminal MATH domain (147 aa). Coding regions of MaMyD88 and MaTRAF6 genomic sequences consisted of five and six exons, respectively. Physicochemical and functional characteristics of the proteins were analysed. Alpha helices were dominant in the secondary structure of the proteins. Homology models of the MaMyD88 and MaTRAF6 domains were constructed applying the comparative modelling method. RT-qPCR was used to analyse the expression of MaMyD88 and MaTRAF6 mRNA transcripts in response to Aeromonas hydrophila challenge. Both genes were highly upregulated in the liver, spleen and kidney during the first 24 h after the challenge. While MyD88 and TRAF6 have been reported in various aquatic species, this is the first report and characterisation of these genes in blunt snout bream. This research also provides evidence of the important roles of these two genes in the blunt snout bream innate immune system.
Figures







Similar articles
-
A novel interleukin-1 receptor-associated kinase 4 from blunt snout bream (Megalobrama amblycephala) is involved in inflammatory response via MyD88-mediated NF-κB signal pathway.Fish Shellfish Immunol. 2022 Aug;127:23-34. doi: 10.1016/j.fsi.2022.05.056. Epub 2022 Jun 1. Fish Shellfish Immunol. 2022. PMID: 35661767
-
Molecular cloning and expression of toll-like receptor 4 (tlr4) in the blunt snout bream (Megalobrama amblycephala).Dev Comp Immunol. 2016 Jun;59:63-76. doi: 10.1016/j.dci.2016.01.009. Epub 2016 Jan 21. Dev Comp Immunol. 2016. PMID: 26802439
-
Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala).Fish Shellfish Immunol. 2013 Aug;35(2):553-63. doi: 10.1016/j.fsi.2013.05.021. Epub 2013 Jun 6. Fish Shellfish Immunol. 2013. PMID: 23748217
-
Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila.Fish Shellfish Immunol. 2015 Jul;45(1):72-82. doi: 10.1016/j.fsi.2015.01.034. Epub 2015 Feb 12. Fish Shellfish Immunol. 2015. PMID: 25681750
-
Cloning, characterization and mRNA expression of interleukin-6 in blunt snout bream (Megalobrama amblycephala).Fish Shellfish Immunol. 2016 Jul;54:639-47. doi: 10.1016/j.fsi.2016.03.005. Epub 2016 Mar 8. Fish Shellfish Immunol. 2016. PMID: 26965748
Cited by
-
Comprehensive Evaluation of Growth Performance, Hematological Parameters, Antioxidant Capacity, Innate Immunity, and Disease Resistance in Crucian Carp (Carassius auratus) Lacking Intermuscular Bones.Antioxidants (Basel). 2025 Apr 8;14(4):443. doi: 10.3390/antiox14040443. Antioxidants (Basel). 2025. PMID: 40298788 Free PMC article.
-
Identification and characterization of the myeloid differentiation factor 88 gene in yellow catfish.3 Biotech. 2018 Oct;8(10):430. doi: 10.1007/s13205-018-1448-z. Epub 2018 Sep 29. 3 Biotech. 2018. PMID: 30305999 Free PMC article.
References
-
- Tort L., Balasch J.C., Mackenzie S. Fish immune system. A crossroads between innate and adaptive responses. Inmunología. 2003;22:277–286.
-
- Basu M., Swai B., Maiti N.K., Routray P., Samanta M. Inductive expression of toll-like receptor 5 (TLR5) and associated downstream signaling molecules following ligand exposure and bacterial infection in the Indian major carp, mrigal (Cirrhinus mrigala) Fish Shellfish Immun. 2012;32:121–131. doi: 10.1016/j.fsi.2011.10.031. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

