Correlation between Reversal of DNA Methylation and Clinical Symptoms in Psoriatic Epidermis Following Narrow-Band UVB Phototherapy
- PMID: 25830654
- PMCID: PMC4580729
- DOI: 10.1038/jid.2015.128
Correlation between Reversal of DNA Methylation and Clinical Symptoms in Psoriatic Epidermis Following Narrow-Band UVB Phototherapy
Abstract
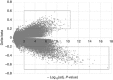
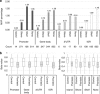
Epigenetic modifications by DNA methylation are associated with a wide range of diseases. Previous studies in psoriasis have concentrated on epigenetic changes in immune cells or in total skin biopsies that include stromal-associated changes. In order to improve our understanding of the role of DNA methylation in psoriasis, we sought to obtain a comprehensive DNA methylation signature specific for the epidermal component of psoriasis and to analyze methylation changes during therapy. Genome-wide DNA methylation profiling of epidermal cells from 12 patients undergoing narrow-band UVB phototherapy and 12 corresponding healthy controls revealed a distinct DNA methylation pattern in psoriasis compared with controls. A total of 3,665 methylation variable positions (MVPs) were identified with an overall hypomethylation in psoriasis patient samples. DNA methylation pattern was reversed at the end of phototherapy in patients showing excellent clinical improvement. Only 7% of phototherapy-affected MVPs (150 out of 2,108) correlate with nearby gene expression. Enrichment of MVPs in enhancers indicates tissue-specific modulation of the transcriptional regulatory machinery in psoriasis. Our study identified key epigenetic events associated with psoriasis pathogenesis and helps understand the dynamic DNA methylation landscape in the human genome.
Figures

Similar articles
-
Epidermal barrier and oxidative stress parameters improve during in 311 nm narrow band UVB phototherapy of plaque type psoriasis.J Dermatol Sci. 2018 Jul;91(1):28-34. doi: 10.1016/j.jdermsci.2018.03.011. Epub 2018 Mar 21. J Dermatol Sci. 2018. PMID: 29610017 Clinical Trial.
-
NOD2 expression in psoriasis before and after treatment with Narrowband Ultraviolet B phototherapy.Arch Dermatol Res. 2025 Jan 16;317(1):260. doi: 10.1007/s00403-024-03690-x. Arch Dermatol Res. 2025. PMID: 39820568
-
Genomic-scale analysis of psoriatic skin reveals differentially expressed insulin-like growth factor-binding protein-7 after phototherapy.Br J Dermatol. 2007 Feb;156(2):289-300. doi: 10.1111/j.1365-2133.2006.07628.x. Br J Dermatol. 2007. PMID: 17223869
-
Phototherapy of Psoriasis, a Chronic Inflammatory Skin Disease.Adv Exp Med Biol. 2017;996:287-294. doi: 10.1007/978-3-319-56017-5_24. Adv Exp Med Biol. 2017. PMID: 29124709 Review.
-
Phosphor for phototherapy: Review on psoriasis.Luminescence. 2017 May;32(3):260-270. doi: 10.1002/bio.3266. Epub 2017 Feb 21. Luminescence. 2017. PMID: 28220603 Review.
Cited by
-
Psoriatic Resolved Skin Epidermal Keratinocytes Retain Disease-Residual Transcriptomic and Epigenomic Profiles.Int J Mol Sci. 2023 Feb 25;24(5):4556. doi: 10.3390/ijms24054556. Int J Mol Sci. 2023. PMID: 36901987 Free PMC article.
-
Loss of the Epigenetic Mark 5-hmC in Psoriasis: Implications for Epidermal Stem Cell Dysregulation.J Invest Dermatol. 2020 Jun;140(6):1266-1275.e3. doi: 10.1016/j.jid.2019.10.016. Epub 2019 Dec 11. J Invest Dermatol. 2020. PMID: 31837302 Free PMC article.
-
Potential evidence for transgenerational epigenetic memory in Arabidopsis thaliana following spaceflight.Commun Biol. 2021 Jul 2;4(1):835. doi: 10.1038/s42003-021-02342-4. Commun Biol. 2021. PMID: 34215844 Free PMC article.
-
Histone Modifications and DNA Methylation in Psoriasis: A Cellular Perspective.Clin Rev Allergy Immunol. 2025 Jan 27;68(1):6. doi: 10.1007/s12016-024-09014-1. Clin Rev Allergy Immunol. 2025. PMID: 39871086 Review.
-
Mergeomics 2.0: a web server for multi-omics data integration to elucidate disease networks and predict therapeutics.Nucleic Acids Res. 2021 Jul 2;49(W1):W375-W387. doi: 10.1093/nar/gkab405. Nucleic Acids Res. 2021. PMID: 34048577 Free PMC article.
References
-
- Bibikova M, Barnes B, Tsan C et al. (2011) High density DNA methylation array with single CpG site resolution. Genomics 98:288–295 - PubMed
-
- Crow JM. (2012) Psoriasis uncovered. Nature 492:S50–S51 - PubMed
-
- Feinberg AP. (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

