Characterization of phospho-(tyrosine)-mimetic calmodulin mutants
- PMID: 25830911
- PMCID: PMC4382182
- DOI: 10.1371/journal.pone.0120798
Characterization of phospho-(tyrosine)-mimetic calmodulin mutants
Abstract
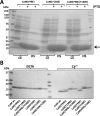
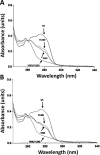
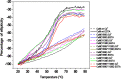
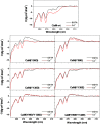
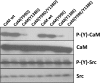
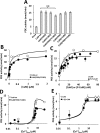
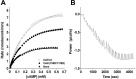
Calmodulin (CaM) phosphorylated at different serine/threonine and tyrosine residues is known to exert differential regulatory effects on a variety of CaM-binding enzymes as compared to non-phosphorylated CaM. In this report we describe the preparation and characterization of a series of phospho-(Y)-mimetic CaM mutants in which either one or the two tyrosine residues present in CaM (Y99 and Y138) were substituted to aspartic acid or glutamic acid. It was expected that the negative charge of the respective carboxyl group of these amino acids mimics the negative charge of phosphate and reproduce the effects that distinct phospho-(Y)-CaM species may have on target proteins. We describe some physicochemical properties of these CaM mutants as compared to wild type CaM, after their expression in Escherichia coli and purification to homogeneity, including: i) changes in their electrophoretic mobility in the absence and presence of Ca2+; ii) ultraviolet (UV) light absorption spectra, far- and near-UV circular dichroism data; iii) thermal stability in the absence and presence of Ca2+; and iv) Tb3+-emitted fluorescence upon tyrosine excitation. We also describe some biochemical properties of these CaM mutants, such as their differential phosphorylation by the tyrosine kinase c-Src, and their action as compared to wild type CaM, on the activity of two CaM-dependent enzymes: cyclic nucleotide phosphodiesterase 1 (PDE1) and endothelial nitric oxide synthase (eNOS) assayed in vitro.
Conflict of interest statement
Figures



Similar articles
-
Characterisation of tyrosine-phosphorylation-defective calmodulin mutants.Protein Expr Purif. 2005 Jun;41(2):384-92. doi: 10.1016/j.pep.2005.01.004. Protein Expr Purif. 2005. PMID: 15866726
-
Tyrosine phosphorylation modulates the interaction of calmodulin with its target proteins.Eur J Biochem. 1999 Jun;262(3):790-802. doi: 10.1046/j.1432-1327.1999.00441.x. Eur J Biochem. 1999. PMID: 10411641
-
Structure and dynamics of calmodulin (CaM) bound to nitric oxide synthase peptides: effects of a phosphomimetic CaM mutation.Biochemistry. 2012 May 1;51(17):3651-61. doi: 10.1021/bi300327z. Epub 2012 Apr 16. Biochemistry. 2012. PMID: 22486744
-
Diversity of calcium action in regulation of mammalian calmodulin-dependent cyclic nucleotide phosphodiesterase.Indian J Biochem Biophys. 2003 Apr;40(2):77-91. Indian J Biochem Biophys. 2003. PMID: 22900295 Review.
-
Phosphorylation of calmodulin. Functional implications.Eur J Biochem. 2002 Aug;269(15):3619-31. doi: 10.1046/j.1432-1033.2002.03038.x. Eur J Biochem. 2002. PMID: 12153558 Review.
Cited by
-
A widespread phage-encoded kinase enables evasion of multiple host antiphage defence systems.Nat Microbiol. 2024 Dec;9(12):3226-3239. doi: 10.1038/s41564-024-01851-2. Epub 2024 Nov 6. Nat Microbiol. 2024. PMID: 39506096
-
Ca2+ Signaling and Src Functions in Tumor Cells.Biomolecules. 2023 Dec 3;13(12):1739. doi: 10.3390/biom13121739. Biomolecules. 2023. PMID: 38136610 Free PMC article. Review.
-
Role of the N-terminal lid in regulating the interaction of phosphorylated MDMX with p53.Oncotarget. 2017 Dec 1;8(68):112825-112840. doi: 10.18632/oncotarget.22829. eCollection 2017 Dec 22. Oncotarget. 2017. PMID: 29348869 Free PMC article.
-
Molecular and biochemical characterization of calmodulin from Echinococcus granulosus.Parasit Vectors. 2017 Dec 4;10(1):597. doi: 10.1186/s13071-017-2545-2. Parasit Vectors. 2017. PMID: 29202858 Free PMC article.
-
Phospho-proteomic discovery of novel signal transducers including thioredoxin-interacting protein as mediators of erythropoietin-dependent human erythropoiesis.Exp Hematol. 2020 Apr;84:29-44. doi: 10.1016/j.exphem.2020.03.003. Epub 2020 Apr 4. Exp Hematol. 2020. PMID: 32259549 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

