The promise of multi-omics and clinical data integration to identify and target personalized healthcare approaches in autism spectrum disorders
- PMID: 25831060
- PMCID: PMC4389910
- DOI: 10.1089/omi.2015.0020
The promise of multi-omics and clinical data integration to identify and target personalized healthcare approaches in autism spectrum disorders
Abstract
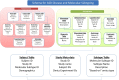
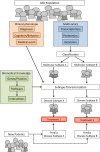
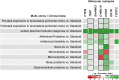
Complex diseases are caused by a combination of genetic and environmental factors, creating a difficult challenge for diagnosis and defining subtypes. This review article describes how distinct disease subtypes can be identified through integration and analysis of clinical and multi-omics data. A broad shift toward molecular subtyping of disease using genetic and omics data has yielded successful results in cancer and other complex diseases. To determine molecular subtypes, patients are first classified by applying clustering methods to different types of omics data, then these results are integrated with clinical data to characterize distinct disease subtypes. An example of this molecular-data-first approach is in research on Autism Spectrum Disorder (ASD), a spectrum of social communication disorders marked by tremendous etiological and phenotypic heterogeneity. In the case of ASD, omics data such as exome sequences and gene and protein expression data are combined with clinical data such as psychometric testing and imaging to enable subtype identification. Novel ASD subtypes have been proposed, such as CHD8, using this molecular subtyping approach. Broader use of molecular subtyping in complex disease research is impeded by data heterogeneity, diversity of standards, and ineffective analysis tools. The future of molecular subtyping for ASD and other complex diseases calls for an integrated resource to identify disease mechanisms, classify new patients, and inform effective treatment options. This in turn will empower and accelerate precision medicine and personalized healthcare.
Figures
References
-
- American Psychiatry Association. (2013). The Diagnostic and Statistical Manual of Mental Disorders: DSM 5 bookpointUS
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical