Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects
- PMID: 25831495
- PMCID: PMC4394309
- DOI: 10.1073/pnas.1502967112
Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects
Abstract
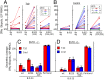
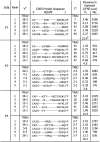
Certain class II MHC (MHCII) alleles in mice and humans confer risk for or protection from type 1 diabetes (T1D). Insulin is a major autoantigen in T1D, but how its peptides are presented to CD4 T cells by MHCII risk alleles has been controversial. In the mouse model of T1D, CD4 T cells respond to insulin B-chain peptide (B:9-23) mimotopes engineered to bind the mouse MHCII molecule, IA(g7), in an unfavorable position or register. Because of the similarities between IA(g7) and human HLA-DQ T1D risk alleles, we examined control and T1D subjects with these risk alleles for CD4 T-cell responses to the same natural B:9-23 peptide and mimotopes. A high proportion of new-onset T1D subjects mounted an inflammatory IFN-γ response much more frequently to one of the mimotope peptides than to the natural peptide. Surprisingly, the control subjects bearing an HLA-DQ risk allele also did. However, these control subjects, especially those with only one HLA-DQ risk allele, very frequently made an IL-10 response, a cytokine associated with regulatory T cells. T1D subjects with established disease also responded to the mimotope rather than the natural B:9-23 peptide in proliferation assays and the proliferating cells were highly enriched in certain T-cell receptor sequences. Our results suggest that the risk of T1D may be related to how an HLA-DQ genotype determines the balance of T-cell inflammatory vs. regulatory responses to insulin, having important implications for the use and monitoring of insulin-specific therapies to prevent diabetes onset.
Keywords: CD4 T cells; autoimmunity; diabetes; insulin; self-tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Harjutsalo V, Sjöberg L, Tuomilehto J. Time trends in the incidence of type 1 diabetes in Finnish children: A cohort study. Lancet. 2008;371(9626):1777–1782. - PubMed
-
- Patterson CC, Dahlquist GG, Gyürüs E, Green A, Soltész G. EURODIAB Study Group Incidence trends for childhood type 1 diabetes in Europe during 1989-2003 and predicted new cases 2005-20: A multicentre prospective registration study. Lancet. 2009;373(9680):2027–2033. - PubMed
-
- Skyler JS. Diabetes Prevention Trial--Type 1 Diabetes Study Group Effects of insulin in relatives of patients with type 1 diabetes mellitus. N Engl J Med. 2002;346(22):1685–1691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 DK080885/DK/NIDDK NIH HHS/United States
- K08DK095995/DK/NIDDK NIH HHS/United States
- R01DK032083/DK/NIDDK NIH HHS/United States
- R01 DK032083/DK/NIDDK NIH HHS/United States
- K08 DK095995/DK/NIDDK NIH HHS/United States
- K99 DK080885/DK/NIDDK NIH HHS/United States
- R01 HL113029/HL/NHLBI NIH HHS/United States
- R01 DK099317/DK/NIDDK NIH HHS/United States
- R00DK080885/DK/NIDDK NIH HHS/United States
- DP3 DK097681/DK/NIDDK NIH HHS/United States
- R01DK099317/DK/NIDDK NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

