The frequency natural antisense transcript first promotes, then represses, frequency gene expression via facultative heterochromatin
- PMID: 25831497
- PMCID: PMC4394252
- DOI: 10.1073/pnas.1406130112
The frequency natural antisense transcript first promotes, then represses, frequency gene expression via facultative heterochromatin
Abstract
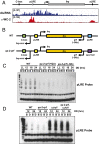
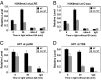
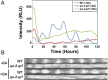
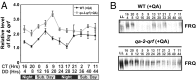
The circadian clock is controlled by a network of interconnected feedback loops that require histone modifications and chromatin remodeling. Long noncoding natural antisense transcripts (NATs) originate from Period in mammals and frequency (frq) in Neurospora. To understand the role of NATs in the clock, we put the frq antisense transcript qrf (frq spelled backwards) under the control of an inducible promoter. Replacing the endogenous qrf promoter altered heterochromatin formation and DNA methylation at frq. In addition, constitutive, low-level induction of qrf caused a dramatic effect on the endogenous rhythm and elevated circadian output. Surprisingly, even though qrf is needed for heterochromatic silencing, induction of qrf initially promoted frq gene expression by creating a more permissible local chromatin environment. The observation that antisense expression can initially promote sense gene expression before silencing via heterochromatin formation at convergent loci is also found when a NAT to hygromycin resistance gene is driven off the endogenous vivid (vvd) promoter in the Δvvd strain. Facultative heterochromatin silencing at frq functions in a parallel pathway to previously characterized VVD-dependent silencing and is needed to establish the appropriate circadian phase. Thus, repression via dicer-independent siRNA-mediated facultative heterochromatin is largely independent of, and occurs alongside, other feedback processes.
Keywords: DNA methylation; circadian rhythm; heterochromatin; natural antisense transcripts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Transcriptional interference by antisense RNA is required for circadian clock function.Nature. 2014 Oct 30;514(7524):650-3. doi: 10.1038/nature13671. Epub 2014 Aug 17. Nature. 2014. PMID: 25132551 Free PMC article.
-
Molecular Regulation of Circadian Chromatin.J Mol Biol. 2020 May 29;432(12):3466-3482. doi: 10.1016/j.jmb.2020.01.009. Epub 2020 Jan 16. J Mol Biol. 2020. PMID: 31954735 Review.
-
Antisense Transcription of the Neurospora Frequency Gene Is Rhythmically Regulated by CSP-1 Repressor but Dispensable for Clock Function.J Biol Rhythms. 2023 Jun;38(3):259-268. doi: 10.1177/07487304231153914. Epub 2023 Mar 1. J Biol Rhythms. 2023. PMID: 36876962 Free PMC article.
-
CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency.PLoS Genet. 2011 Jul;7(7):e1002166. doi: 10.1371/journal.pgen.1002166. Epub 2011 Jul 21. PLoS Genet. 2011. PMID: 21811413 Free PMC article.
-
The neurospora circadian system.J Biol Rhythms. 2004 Oct;19(5):414-24. doi: 10.1177/0748730404269116. J Biol Rhythms. 2004. PMID: 15534321 Review.
Cited by
-
The circadian system as an organizer of metabolism.Fungal Genet Biol. 2016 May;90:39-43. doi: 10.1016/j.fgb.2015.10.002. Epub 2015 Oct 20. Fungal Genet Biol. 2016. PMID: 26498192 Free PMC article. Review.
-
Natural Variation of the Circadian Clock in Neurospora.Adv Genet. 2017;99:1-37. doi: 10.1016/bs.adgen.2017.09.001. Epub 2017 Oct 12. Adv Genet. 2017. PMID: 29050553 Free PMC article. Review.
-
Timing without coding: How do long non-coding RNAs regulate circadian rhythms?Semin Cell Dev Biol. 2022 Jun;126:79-86. doi: 10.1016/j.semcdb.2021.04.020. Epub 2021 Jun 9. Semin Cell Dev Biol. 2022. PMID: 34116930 Free PMC article. Review.
-
Developmental Dynamics of Long Noncoding RNA Expression during Sexual Fruiting Body Formation in Fusarium graminearum.mBio. 2018 Aug 14;9(4):e01292-18. doi: 10.1128/mBio.01292-18. mBio. 2018. PMID: 30108170 Free PMC article.
-
Transcriptome analysis of smut fungi reveals widespread intergenic transcription and conserved antisense transcript expression.BMC Genomics. 2017 May 2;18(1):340. doi: 10.1186/s12864-017-3720-8. BMC Genomics. 2017. PMID: 28464849 Free PMC article.
References
-
- Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. - PubMed
-
- Brunner M, Káldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 2008;68(2):255–262. - PubMed
-
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276(5313):763–769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

