Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis
- PMID: 25831531
- PMCID: PMC4394283
- DOI: 10.1073/pnas.1421504112
Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis
Abstract
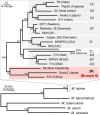
Mycobacterium lepromatosis is an uncultured human pathogen associated with diffuse lepromatous leprosy and a reactional state known as Lucio's phenomenon. By using deep sequencing with and without DNA enrichment, we obtained the near-complete genome sequence of M. lepromatosis present in a skin biopsy from a Mexican patient, and compared it with that of Mycobacterium leprae, which has undergone extensive reductive evolution. The genomes display extensive synteny and are similar in size (∼3.27 Mb). Protein-coding genes share 93% nucleotide sequence identity, whereas pseudogenes are only 82% identical. The events that led to pseudogenization of 50% of the genome likely occurred before divergence from their most recent common ancestor (MRCA), and both M. lepromatosis and M. leprae have since accumulated new pseudogenes or acquired specific deletions. Functional comparisons suggest that M. lepromatosis has lost several enzymes required for amino acid synthesis whereas M. leprae has a defective heme pathway. M. lepromatosis has retained all functions required to infect the Schwann cells of the peripheral nervous system and therefore may also be neuropathogenic. A phylogeographic survey of 227 leprosy biopsies by differential PCR revealed that 221 contained M. leprae whereas only six, all from Mexico, harbored M. lepromatosis. Phylogenetic comparisons indicate that M. lepromatosis is closer than M. leprae to the MRCA, and a Bayesian dating analysis suggests that they diverged from their MRCA approximately 13.9 Mya. Thus, despite their ancient separation, the two leprosy bacilli are remarkably conserved and still cause similar pathologic conditions.
Keywords: Mycobacterium leprae; Mycobacterium lepromatosis; comparative genomics; genome sequencing; reductive evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization Global leprosy: Update on the 2012 situation. Wkly Epidemiol Rec. 2013;88(35):365–379. - PubMed
-
- Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. - PubMed
-
- Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363(9416):1209–1219. - PubMed
-
- Lucio R, Alsarado Y. Opusculo sobre el mal de San Làzaro, ó elefanciasis de los griegos, escrito por los profesores de medicina y cirugia. Imprento de M. Murguia y Compañia; Mexico City, Mexico: 1852.
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP047206
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

