Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment
- PMID: 25831549
- PMCID: PMC4385470
- DOI: 10.1126/science.1258699
Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment
Abstract
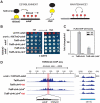
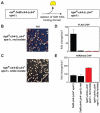
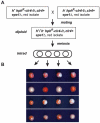
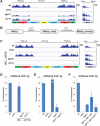
Changes in histone posttranslational modifications are associated with epigenetic states that define distinct patterns of gene expression. It remains unclear whether epigenetic information can be transmitted through histone modifications independently of specific DNA sequence, DNA methylation, or RNA interference. Here we show that, in the fission yeast Schizosaccharomyces pombe, ectopically induced domains of histone H3 lysine 9 methylation (H3K9me), a conserved marker of heterochromatin, are inherited through several mitotic and meiotic cell divisions after removal of the sequence-specific initiator. The putative JmjC domain H3K9 demethylase, Epe1, and the chromodomain of the H3K9 methyltransferase, Clr4/Suv39h, play opposing roles in maintaining silent H3K9me domains. These results demonstrate how a direct "read-write" mechanism involving Clr4 propagates histone modifications and allows histones to act as carriers of epigenetic information.
Copyright © 2015, American Association for the Advancement of Science.
Figures



References
-
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trlthorax group proteins. Annu. Rev. Genet. 2004;38:413–443. Medline doi:10.1146/annurev.genet.38.072902.091907. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251260. Medline doi:10.1038/38444. - PubMed
-
- Jenuwein T, Allls CD. Translating the histone code. Science. 2001;293:1074–1080. Medline doi:10.1126/science.1063127. - PubMed
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. Medline doi:10.1016/j.cell.2007.01.015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

