Characterization of an α-l-fucosidase from the periodontal pathogen Tannerella forsythia
- PMID: 25831954
- PMCID: PMC4413431
- DOI: 10.1080/21505594.2015.1010982
Characterization of an α-l-fucosidase from the periodontal pathogen Tannerella forsythia
Abstract
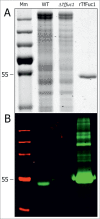
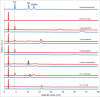
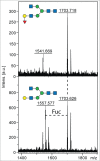
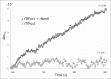
The periodontal pathogen Tannerella forsythia expresses several glycosidases which are linked to specific growth requirements and are involved in the invasion of host tissues. α-l-Fucosyl residues are exposed on various host glycoconjugates and, thus, the α-l-fucosidases predicted in the T. forsythia ATCC 43037 genome could potentially serve roles in host-pathogen interactions. We describe the molecular cloning and characterization of the putative fucosidase TfFuc1 (encoded by the bfo_2737 = Tffuc1 gene), previously reported to be present in an outer membrane preparation. In terms of sequence, this 51-kDa protein is a member of the glycosyl hydrolase family GH29. Using an artificial substrate, p-nitrophenyl-α-fucose (KM 670 μM), the enzyme was determined to have a pH optimum of 9.0 and to be competitively inhibited by fucose and deoxyfuconojirimycin. TfFuc1 was shown here to be a unique α(1,2)-fucosidase that also possesses α(1,6) specificity on small unbranched substrates. It is active on mucin after sialidase-catalyzed removal of terminal sialic acid residues and also removes fucose from blood group H. Following knock-out of the Tffuc1 gene and analyzing biofilm formation and cell invasion/adhesion of the mutant in comparison to the wild-type, it is most likely that the enzyme does not act extracellularly. Biochemically interesting as the first fucosidase in T. forsythia to be characterized, the biological role of TfFuc1 may well be in the metabolism of short oligosaccharides in the periplasm, thereby indirectly contributing to the virulence of this organism. TfFuc1 is the first glycosyl hydrolase in the GH29 family reported to be a specific α(1,2)-fucosidase.
Keywords: 2) fucosidase; 4-nitrophenyl-α-l-fucopyranoside; Amp, ampicillin; BHI, brain heart infusion medium; CBB, Coomassie brilliant blue G 250; DFJ, deoxyfuconojirimycin; Erm, erythromycin; FDH, fucose dehydrogenase; HPAEC, high-performance anion-exchange chromatography with pulsed amperometric detection; LC-ESI-MS, liquid chromatography-electrospray ionisation-mass spectrometry; NAM, N-acetylmuramic acid; PBS, phosphate-buffered saline; SDS-PAGE, sodium dodecylsulphate polyacrylamide gel electrophoresis; T. forsythia, Tannerella forsythia ATCC 43037; TfFuc1, T. forsythia ATCC 43037 fucosidase-1 encoded by the bfo_2737 gene, equally Tffuc1; WT, wild-type bacterium.; enzyme activity; enzyme specificity; oral pathogen; pNP-fucose; periodontitis; rTfFuc-1, recombinant TfFuc1 enzyme; tannerella forsythia; α(1.
Figures


Similar articles
-
Infant Gut Microbial Metagenome Mining of α-l-Fucosidases with Activity on Fucosylated Human Milk Oligosaccharides and Glycoconjugates.Microbiol Spectr. 2022 Aug 31;10(4):e0177522. doi: 10.1128/spectrum.01775-22. Epub 2022 Aug 9. Microbiol Spectr. 2022. PMID: 35943155 Free PMC article.
-
Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates.Glycobiology. 2009 Sep;19(9):1010-7. doi: 10.1093/glycob/cwp082. Epub 2009 Jun 11. Glycobiology. 2009. PMID: 19520709
-
The fucosidase-pool of Emticicia oligotrophica: Biochemical characterization and transfucosylation potential.Glycobiology. 2016 Aug;26(8):871-879. doi: 10.1093/glycob/cww030. Epub 2016 Mar 3. Glycobiology. 2016. PMID: 26941394
-
Structure and function of microbial α-l-fucosidases: a mini review.Essays Biochem. 2023 Apr 18;67(3):399-414. doi: 10.1042/EBC20220158. Essays Biochem. 2023. PMID: 36805644 Free PMC article. Review.
-
Mammalian alpha-L-fucosidases.Comp Biochem Physiol B. 1991;99(3):479-88. doi: 10.1016/0305-0491(91)90327-a. Comp Biochem Physiol B. 1991. PMID: 1769200 Review.
Cited by
-
Identification and Characterization of a Novel α-L-Fucosidase from Enterococcus gallinarum and Its Application for Production of 2'-Fucosyllactose.Int J Mol Sci. 2023 Jul 17;24(14):11555. doi: 10.3390/ijms241411555. Int J Mol Sci. 2023. PMID: 37511315 Free PMC article.
-
Tannerella forsythia strains display different cell-surface nonulosonic acids: biosynthetic pathway characterization and first insight into biological implications.Glycobiology. 2017 Apr 1;27(4):342-357. doi: 10.1093/glycob/cww129. Glycobiology. 2017. PMID: 27986835 Free PMC article.
-
A Robust α-l-Fucosidase from Prevotella nigrescens for Glycoengineering Therapeutic Antibodies.ACS Chem Biol. 2024 Jul 19;19(7):1515-1524. doi: 10.1021/acschembio.4c00196. Epub 2024 Jun 24. ACS Chem Biol. 2024. PMID: 38912881 Free PMC article.
-
A Combination of Structural, Genetic, Phenotypic and Enzymatic Analyses Reveals the Importance of a Predicted Fucosyltransferase to Protein O-Glycosylation in the Bacteroidetes.Biomolecules. 2021 Nov 30;11(12):1795. doi: 10.3390/biom11121795. Biomolecules. 2021. PMID: 34944439 Free PMC article.
-
Nonulosonic acids contribute to the pathogenicity of the oral bacterium Tannerella forsythia.Interface Focus. 2019 Apr 6;9(2):20180064. doi: 10.1098/rsfs.2018.0064. Epub 2019 Feb 15. Interface Focus. 2019. PMID: 30842870 Free PMC article. Review.
References
-
- Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V., et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the global burden of disease study 2010. Lancet 2012; 380:2163-96; PMID:23245607; http://dx.doi.org/10.1016/S0140-6736(12)61729-2 - DOI - PMC - PubMed
-
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL, Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25:134-44; PMID:9495612; http://dx.doi.org/10.1111/j.1600-051X.1998.tb02419.x - DOI - PubMed
-
- Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol 2008; 23:196-205; PMID:18402605; http://dx.doi.org/10.1111/j.1399-302X.2007.00411.x - DOI - PubMed
-
- Cullinan MP, Ford PJ, Seymour GJ. Periodontal disease and systemic health: current status. Aust Dent J 2009; 54:S62-9; PMID:19737269; http://dx.doi.org/10.1111/j.1834-7819.2009.01144.x - DOI - PubMed
-
- Posch G, Sekot G, Friedrich V, Megson ZA, Koerdt A, Messner P, Schäffer C. Glycobiology aspects of the periodontal pathogen tannerella forsythia. Biomolecules 2012; 2:467-82; PMID:24970146; http://dx.doi.org/10.3390/biom2040467 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
