Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014
- PMID: 25833052
- PMCID: PMC4474294
- DOI: 10.1128/JVI.00459-15
Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014
Abstract
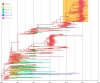
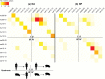
The diversity of influenza A viruses in swine (swIAVs) presents an important pandemic threat. Knowledge of the human-swine interface is particularly important for understanding how viruses with pandemic potential evolve in swine hosts. Through phylogenetic analysis of contemporary swIAVs in the United States, we demonstrate that human-to-swine transmission of pandemic H1N1 (pH1N1) viruses has occurred continuously in the years following the 2009 H1N1 pandemic and has been an important contributor to the genetic diversity of U.S. swIAVs. Although pandemic H1 and N1 segments had been largely removed from the U.S. swine population by 2013 via reassortment with other swIAVs, these antigens reemerged following multiple human-to-swine transmission events during the 2013-2014 seasonal epidemic. These findings indicate that the six internal gene segments from pH1N1 viruses are likely to be sustained long term in the U.S. swine population, with periodic reemergence of pandemic hemagglutinin (HA) and neuraminidase (NA) segments in association with seasonal pH1N1 epidemics in humans. Vaccinating U.S. swine workers may reduce infection of both humans and swine and in turn limit the role of humans as sources of influenza virus diversity in pigs.
Importance: Swine are important hosts in the evolution of influenza A viruses with pandemic potential. Here, we analyze influenza virus sequence data generated by the U.S. Department of Agriculture's national surveillance system to identify the central role of humans in the reemergence of pandemic H1N1 (pH1N1) influenza viruses in U.S. swine herds in 2014. These findings emphasize the important role of humans as continuous sources of influenza virus diversity in swine and indicate that influenza viruses with pandemic HA and NA segments are likely to continue to reemerge in U.S. swine in association with seasonal pH1N1 epidemics in humans.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Scholtissek C. 1990. Pigs as the “mixing vessel” for the creation of new pandemic influenza A viruses. Med Princ Pract 2:65–71.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

