Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner
- PMID: 25833711
- PMCID: PMC4472013
- DOI: 10.1091/mbc.E14-12-1577
Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner
Abstract
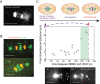
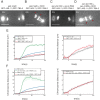
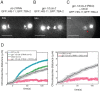
During mitosis, chromosomes are connected to a microtubule-based spindle. Current models propose that displacement of the spindle poles and/or the activity of kinetochore microtubules generate mechanical forces that segregate sister chromatids. Using laser destruction of the centrosomes during Caenorhabditis elegans mitosis, we show that neither of these mechanisms is necessary to achieve proper chromatid segregation. Our results strongly suggest that an outward force generated by the spindle midzone, independently of centrosomes, is sufficient to segregate chromosomes in mitotic cells. Using mutant and RNAi analysis, we show that the microtubule-bundling protein SPD-1/MAP-65 and BMK-1/kinesin-5 act as a brake opposing the force generated by the spindle midzone. Conversely, we identify a novel role for two microtubule-growth and nucleation agents, Ran and CLASP, in the establishment of the centrosome-independent force during anaphase. Their involvement raises the interesting possibility that microtubule polymerization of midzone microtubules is continuously required to sustain chromosome segregation during mitosis.
© 2015 Nahaboo et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures



Similar articles
-
Kinetochore Recruitment of the Spindle and Kinetochore-Associated (Ska) Complex Is Regulated by Centrosomal PP2A in Caenorhabditis elegans.Genetics. 2019 Jun;212(2):509-522. doi: 10.1534/genetics.119.302105. Epub 2019 Apr 24. Genetics. 2019. PMID: 31018924 Free PMC article.
-
BUB-1 promotes amphitelic chromosome biorientation via multiple activities at the kinetochore.Elife. 2018 Dec 14;7:e40690. doi: 10.7554/eLife.40690. Elife. 2018. PMID: 30547880 Free PMC article.
-
The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles.Mol Biol Cell. 2005 Feb;16(2):742-56. doi: 10.1091/mbc.e04-08-0682. Epub 2004 Nov 17. Mol Biol Cell. 2005. PMID: 15548597 Free PMC article.
-
Factors that Control Mitotic Spindle Dynamics.Adv Exp Med Biol. 2017;925:89-101. doi: 10.1007/5584_2016_74. Adv Exp Med Biol. 2017. PMID: 27722958 Review.
-
Centrosomes and the art of mitotic spindle maintenance.Int Rev Cell Mol Biol. 2014;313:179-217. doi: 10.1016/B978-0-12-800177-6.00006-2. Int Rev Cell Mol Biol. 2014. PMID: 25376493 Review.
Cited by
-
Anaphase B.Biology (Basel). 2016 Dec 8;5(4):51. doi: 10.3390/biology5040051. Biology (Basel). 2016. PMID: 27941648 Free PMC article. Review.
-
How Essential Kinesin-5 Becomes Non-Essential in Fission Yeast: Force Balance and Microtubule Dynamics Matter.Cells. 2020 May 7;9(5):1154. doi: 10.3390/cells9051154. Cells. 2020. PMID: 32392819 Free PMC article. Review.
-
Chromosome segregation occurs by microtubule pushing in oocytes.Nat Commun. 2017 Nov 14;8(1):1499. doi: 10.1038/s41467-017-01539-8. Nat Commun. 2017. PMID: 29133801 Free PMC article.
-
New alleles of C. elegans gene cls-2 (R107.6), called xc3, xc4, and xc5.MicroPubl Biol. 2017 Dec 19;2017:10.17912/W2RQ2X. doi: 10.17912/W2RQ2X. MicroPubl Biol. 2017. PMID: 32550356 Free PMC article. No abstract available.
-
Microtubule Sliding within the Bridging Fiber Pushes Kinetochore Fibers Apart to Segregate Chromosomes.Dev Cell. 2017 Oct 9;43(1):11-23.e6. doi: 10.1016/j.devcel.2017.09.010. Dev Cell. 2017. PMID: 29017027 Free PMC article.
References
-
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

