Avoidable errors in the modelling of outbreaks of emerging pathogens, with special reference to Ebola
- PMID: 25833863
- PMCID: PMC4426634
- DOI: 10.1098/rspb.2015.0347
Avoidable errors in the modelling of outbreaks of emerging pathogens, with special reference to Ebola
Abstract
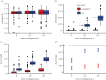
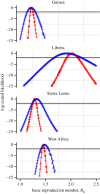
As an emergent infectious disease outbreak unfolds, public health response is reliant on information on key epidemiological quantities, such as transmission potential and serial interval. Increasingly, transmission models fit to incidence data are used to estimate these parameters and guide policy. Some widely used modelling practices lead to potentially large errors in parameter estimates and, consequently, errors in model-based forecasts. Even more worryingly, in such situations, confidence in parameter estimates and forecasts can itself be far overestimated, leading to the potential for large errors that mask their own presence. Fortunately, straightforward and computationally inexpensive alternatives exist that avoid these problems. Here, we first use a simulation study to demonstrate potential pitfalls of the standard practice of fitting deterministic models to cumulative incidence data. Next, we demonstrate an alternative based on stochastic models fit to raw data from an early phase of 2014 West Africa Ebola virus disease outbreak. We show not only that bias is thereby reduced, but that uncertainty in estimates and forecasts is better quantified and that, critically, lack of model fit is more readily diagnosed. We conclude with a short list of principles to guide the modelling response to future infectious disease outbreaks.
Keywords: Ebola virus disease; emerging infectious disease; forecast.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
