The N-terminal Set-β Protein Isoform Induces Neuronal Death
- PMID: 25833944
- PMCID: PMC4505589
- DOI: 10.1074/jbc.M114.633883
The N-terminal Set-β Protein Isoform Induces Neuronal Death
Abstract
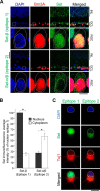
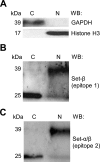
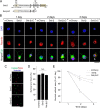
Set-β protein plays different roles in neurons, but the diversity of Set-β neuronal isoforms and their functions have not been characterized. The expression and subcellular localization of Set-β are altered in Alzheimer disease, cleavage of Set-β leads to neuronal death after stroke, and the full-length Set-β regulates retinal ganglion cell (RGC) and hippocampal neuron axon growth and regeneration in a subcellular localization-dependent manner. Here we used various biochemical approaches to investigate Set-β isoforms and their role in the CNS, using the same type of neurons, RGCs, across studies. We found multiple alternatively spliced isoforms expressed from the Set locus in purified RGCs. Set transcripts containing the Set-β-specific exon were the most highly expressed isoforms. We also identified a novel, alternatively spliced Set-β transcript lacking the nuclear localization signal and demonstrated that the full-length (∼39-kDa) Set-β is localized predominantly in the nucleus, whereas a shorter (∼25-kDa) Set-β isoform is localized predominantly in the cytoplasm. Finally, we show that an N-terminal Set-β cleavage product can induce neuronal death.
Keywords: Set-β; alternative splicing; cell death; neuron; protein translocation; subcellular fractionation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Seo S. B., McNamara P., Heo S., Turner A., Lane W. S., Chakravarti D. (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104, 119–130 - PubMed
-
- Gamble M. J., Fisher R. P. (2007) SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 14, 548–555 - PubMed
-
- De Koning L., Corpet A., Haber J. E., Almouzni G. (2007) Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14, 997–1007 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

