Protective Efficacy of the Conserved NP, PB1, and M1 Proteins as Immunogens in DNA- and Vaccinia Virus-Based Universal Influenza A Virus Vaccines in Mice
- PMID: 25834017
- PMCID: PMC4446406
- DOI: 10.1128/CVI.00091-15
Protective Efficacy of the Conserved NP, PB1, and M1 Proteins as Immunogens in DNA- and Vaccinia Virus-Based Universal Influenza A Virus Vaccines in Mice
Abstract
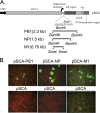
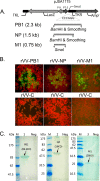
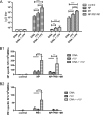
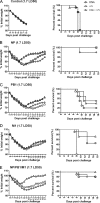
The conventional hemagglutinin (HA)- and neuraminidase (NA)-based influenza vaccines need to be updated most years and are ineffective if the glycoprotein HA of the vaccine strains is a mismatch with that of the epidemic strain. Universal vaccines targeting conserved viral components might provide cross-protection and thus complement and improve conventional vaccines. In this study, we generated DNA plasmids and recombinant vaccinia viruses expressing the conserved proteins nucleoprotein (NP), polymerase basic 1 (PB1), and matrix 1 (M1) from influenza virus strain A/Beijing/30/95 (H3N2). BALB/c mice were immunized intramuscularly with a single vaccine based on NP, PB1, or M1 alone or a combination vaccine based on all three antigens and were then challenged with lethal doses of the heterologous influenza virus strain A/PR/8/34 (H1N1). Vaccines based on NP, PB1, and M1 provided complete or partial protection against challenge with 1.7 50% lethal dose (LD50) of PR8 in mice. Of the three antigens, NP-based vaccines induced protection against 5 LD50 and 10 LD50 and thus exhibited the greatest protective effect. Universal influenza vaccines based on the combination of NP, PB1, and M1 induced a strong immune response and thus might be an alternative approach to addressing future influenza virus pandemics.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Co-administration of certain DNA vaccine combinations expressing different H5N1 influenza virus antigens can be beneficial or detrimental to immune protection.Vaccine. 2012 Jan 11;30(3):626-36. doi: 10.1016/j.vaccine.2011.11.017. Epub 2011 Nov 23. Vaccine. 2012. PMID: 22119588
-
Immunization with influenza A NP-expressing vaccinia virus recombinant protects mice against experimental infection with human and avian influenza viruses.Arch Virol. 2006 May;151(5):921-31. doi: 10.1007/s00705-005-0676-9. Epub 2005 Nov 15. Arch Virol. 2006. PMID: 16292596
-
Incorporation of conserved nucleoprotein into influenza virus-like particles could provoke a broad protective immune response in BALB/c mice and chickens.Virus Res. 2015 Jan 2;195:35-42. doi: 10.1016/j.virusres.2014.09.018. Virus Res. 2015. PMID: 25312452
-
Progress on adenovirus-vectored universal influenza vaccines.Hum Vaccin Immunother. 2015;11(5):1209-22. doi: 10.1080/21645515.2015.1016674. Hum Vaccin Immunother. 2015. PMID: 25876176 Free PMC article. Review.
-
Identification of effective constituents of influenza vaccine by immunization with plasmid DNAs encoding viral proteins.Jpn J Infect Dis. 2000 Dec;53(6):219-28. Jpn J Infect Dis. 2000. PMID: 11227019 Review.
Cited by
-
Improving Cross-Protection against Influenza Virus Using Recombinant Vaccinia Vaccine Expressing NP and M2 Ectodomain Tandem Repeats.Virol Sin. 2019 Oct;34(5):583-591. doi: 10.1007/s12250-019-00138-9. Epub 2019 Jun 25. Virol Sin. 2019. PMID: 31240620 Free PMC article.
-
The Next Generation of Influenza Vaccines: Towards a Universal Solution.Vaccines (Basel). 2021 Jan 7;9(1):26. doi: 10.3390/vaccines9010026. Vaccines (Basel). 2021. PMID: 33430278 Free PMC article. Review.
-
Protective immunity against influenza in HLA-A2 transgenic mice by modified vaccinia virus Ankara vectored vaccines containing internal influenza proteins.Pathog Glob Health. 2017 Mar;111(2):76-82. doi: 10.1080/20477724.2016.1275465. Epub 2017 Jan 12. Pathog Glob Health. 2017. PMID: 28079473 Free PMC article.
-
Recalling the Future: Immunological Memory Toward Unpredictable Influenza Viruses.Front Immunol. 2019 Jul 2;10:1400. doi: 10.3389/fimmu.2019.01400. eCollection 2019. Front Immunol. 2019. PMID: 31312199 Free PMC article. Review.
-
Impact of Individual Viral Gene Segments from Influenza A/H5N8 Virus on the Protective Efficacy of Inactivated Subtype-Specific Influenza Vaccine.Pathogens. 2021 Mar 19;10(3):368. doi: 10.3390/pathogens10030368. Pathogens. 2021. PMID: 33808583 Free PMC article.
References
-
- Manzoli L, Schioppa F, Boccia A, Villari P. 2007. The efficacy of influenza vaccine for healthy children: a meta-analysis evaluating potential sources of variation in efficacy estimates including study quality. Pediatr Infect Dis J 26:97–106. doi:10.1097/01.inf.0000253053.01151.bd. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

