Lipin-1 regulates cancer cell phenotype and is a potential target to potentiate rapamycin treatment
- PMID: 25834103
- PMCID: PMC4484455
- DOI: 10.18632/oncotarget.3595
Lipin-1 regulates cancer cell phenotype and is a potential target to potentiate rapamycin treatment
Abstract
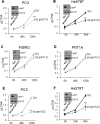
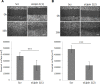
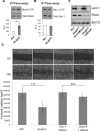
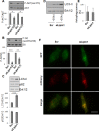
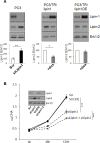
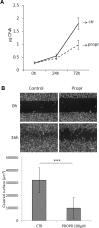
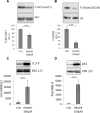
Lipogenesis inhibition was reported to induce apoptosis and repress proliferation of cancer cells while barely affecting normal cells. Lipins exhibit dual function as enzymes catalyzing the dephosphorylation of phosphatidic acid to diacylglycerol and as co-transcriptional regulators. Thus, they are able to regulate lipid homeostasis at several nodal points. Here, we show that lipin-1 is up-regulated in several cancer cell lines and overexpressed in 50 % of high grade prostate cancers. The proliferation of prostate and breast cancer cells, but not of non-tumorigenic cells, was repressed upon lipin-1 knock-down. Lipin-1 depletion also decreased cancer cell migration through RhoA activation. Lipin-1 silencing did not significantly affect global lipid synthesis but enhanced the cellular concentration of phosphatidic acid. In parallel, autophagy was induced while AKT and ribosomal protein S6 phosphorylation were repressed. We also observed a compensatory regulation between lipin-1 and lipin-2 and demonstrated that their co-silencing aggravates the phenotype induced by lipin-1 silencing alone. Most interestingly, lipin-1 depletion or lipins inhibition with propranolol sensitized cancer cells to rapamycin. These data indicate that lipin-1 controls main cellular processes involved in cancer progression and that its targeting, alone or in combination with other treatments, could open new avenues in anticancer therapy.
Keywords: RhoA; lipin-1; metabolism; prostate cancer; rapamycin.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

