Analysis of RNA Interference Lines Identifies New Functions of Maternally-Expressed Genes Involved in Embryonic Patterning in Drosophila melanogaster
- PMID: 25834215
- PMCID: PMC4478533
- DOI: 10.1534/g3.115.017517
Analysis of RNA Interference Lines Identifies New Functions of Maternally-Expressed Genes Involved in Embryonic Patterning in Drosophila melanogaster
Abstract
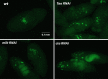
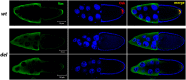
Embryonic patterning in Drosophila melanogaster is initially established through the activity of a number of maternally expressed genes that are expressed during oogenesis. mRNAs from some of these genes accumulate in the posterior pole plasm of the oocyte and early embryo and localize further into RNA islands, which are transient ring-like structures that form around the nuclei of future primordial germ cells (pole cells) at stage 3 of embryogenesis. As mRNAs from several genes with known functions in anterior-posterior patterning and/or germ cell specification accumulate in RNA islands, we hypothesized that some other mRNAs that localize in this manner might also function in these developmental processes. To test this, we investigated the developmental functions of 51 genes whose mRNAs accumulate in RNA islands by abrogating their activity in the female germline using RNA interference. This analysis revealed requirements for ttk, pbl, Hip14, eIF5, eIF4G, and CG9977 for progression through early oogenesis. We observed dorsal appendage defects in a proportion of eggs produced by females expressing double-stranded RNA targeting Mkrn1 or jvl, implicating these two genes in dorsal-ventral patterning. In addition, posterior patterning defects and a reduction in pole cell number were seen in the progeny of Mkrn1 females. Because the mammalian ortholog of Mkrn1 acts as an E3 ubiquitin ligase, these results suggest an additional link between protein ubiquitination and pole plasm activity.
Keywords: embryonic patterning; germ plasm; localized mRNAs; oogenesis; pole cells.
Copyright © 2015 Liu and Lasko.
Figures





Similar articles
-
Multiple Functions of the DEAD-Box Helicase Vasa in Drosophila Oogenesis.Results Probl Cell Differ. 2017;63:127-147. doi: 10.1007/978-3-319-60855-6_6. Results Probl Cell Differ. 2017. PMID: 28779316 Review.
-
The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo.Development. 2009 Jan;136(1):95-105. doi: 10.1242/dev.027698. Epub 2008 Nov 26. Development. 2009. PMID: 19036801
-
Clathrin heavy chain plays multiple roles in polarizing the Drosophila oocyte downstream of Bic-D.Development. 2014 May;141(9):1915-26. doi: 10.1242/dev.099432. Epub 2014 Apr 9. Development. 2014. PMID: 24718986
-
Molecular movements in oocyte patterning and pole cell differentiation.Bioessays. 1992 Aug;14(8):507-12. doi: 10.1002/bies.950140802. Bioessays. 1992. PMID: 1365903 Review.
-
C-terminal residues specific to Vasa among DEAD-box helicases are required for its functions in piRNA biogenesis and embryonic patterning.Dev Genes Evol. 2016 Nov;226(6):401-412. doi: 10.1007/s00427-016-0560-5. Epub 2016 Aug 29. Dev Genes Evol. 2016. PMID: 27572922
Cited by
-
Knock down analysis reveals critical phases for specific oskar noncoding RNA functions during Drosophila oogenesis.G3 (Bethesda). 2021 Dec 8;11(12):jkab340. doi: 10.1093/g3journal/jkab340. G3 (Bethesda). 2021. PMID: 34586387 Free PMC article.
-
The Dynamic Regulation of mRNA Translation and Ribosome Biogenesis During Germ Cell Development and Reproductive Aging.Front Cell Dev Biol. 2021 Nov 3;9:710186. doi: 10.3389/fcell.2021.710186. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34805139 Free PMC article. Review.
-
Makorin 1 controls embryonic patterning by alleviating Bruno1-mediated repression of oskar translation.PLoS Genet. 2020 Jan 24;16(1):e1008581. doi: 10.1371/journal.pgen.1008581. eCollection 2020 Jan. PLoS Genet. 2020. PMID: 31978041 Free PMC article.
-
The Transgenic RNAi Project at Harvard Medical School: Resources and Validation.Genetics. 2015 Nov;201(3):843-52. doi: 10.1534/genetics.115.180208. Epub 2015 Aug 28. Genetics. 2015. PMID: 26320097 Free PMC article.
References
-
- Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S., 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1: 431–437. - PubMed
-
- Chicoine J., Benoit P., Gamberi C., Paliouras M., Simonelig M., et al. , 2007. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell 13: 691–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
