Termination of DNA replication forks: "Breaking up is hard to do"
- PMID: 25835602
- PMCID: PMC4615769
- DOI: 10.1080/19491034.2015.1035843
Termination of DNA replication forks: "Breaking up is hard to do"
Abstract
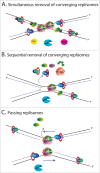
To ensure duplication of the entire genome, eukaryotic DNA replication initiates from thousands of replication origins. The replication forks move through the chromatin until they encounter forks from neighboring origins. During replication fork termination forks converge, the replisomes disassemble and topoisomerase II resolves the daughter DNA molecules. If not resolved efficiently, terminating forks result in genomic instability through the formation of pathogenic structures. Our recent findings shed light onto the mechanism of replisome disassembly upon replication fork termination. We have shown that termination-specific polyubiquitylation of the replicative helicase component - Mcm7, leads to dissolution of the active helicase in a process dependent on the p97/VCP/Cdc48 segregase. The inhibition of terminating helicase disassembly resulted in a replication termination defect. In this extended view we present hypothetical models of replication fork termination and discuss remaining and emerging questions in the DNA replication termination field.
Keywords: CMG, Cdc45, Mcm2–7, GINS complex; CRL, cullin-RING ligase; D loop, displacement loop; DDR, DNA damage response; DNA replication; DSB, double strand break; DUB, deubiquitylating enzyme; ER, endoplasmic reticulum; ERAD, endoplasmic reticulum associated protein degradation; GINS, Go-Ichi-Ni-San, complex made of Sld5, Psf1, Psf2, Psf3; ICL, intra-strand crosslink; MCM, Minichromosome maintenance; Mcm2–7; OriC, chromosomal replication origin; R loop, RNA:DNA hybrid; RING, really interesting gene; RPC, Replisome Progression Complex; Ter, termination site; Tus-Ter, terminus utilisation substance - termination; Xenopus; p97 segregase; replication termination; replicative helicase; replisome; ubiquitin.
Figures


References
-
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953. Apr 25; 171(4356):737-8; PMID:13054692; http://dx.doi.org/10.1038/171737a0 - DOI - PubMed
-
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953. May 30; 171(4361):964-7; PMID:13063483; http://dx.doi.org/10.1038/171964b0 - DOI - PubMed
-
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 2010. Aug 27; 39(4):595-605; PMID:20797631; http://dx.doi.org/10.1016/j.molcel.2010.07.024 - DOI - PMC - PubMed
-
- Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. Avoiding chromosome pathology when replication forks collide. Nature 2013. Jul 28; 500(7464):608-11; PMID:23892781; http://dx.doi.org/10.1038/nature12312 - DOI - PMC - PubMed
-
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 2014. Oct 24; 346(6208):477-81; PMID:25342805; http://dx.doi.org/10.1126/science.1253585 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
