Hantaan virus infection induces both Th1 and ThGranzyme B+ cell immune responses that associated with viral control and clinical outcome in humans
- PMID: 25836633
- PMCID: PMC4383613
- DOI: 10.1371/journal.ppat.1004788
Hantaan virus infection induces both Th1 and ThGranzyme B+ cell immune responses that associated with viral control and clinical outcome in humans
Abstract
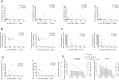
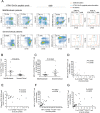
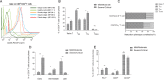
Hantaviruses infection causing severe emerging diseases with high mortality rates in humans has become public health concern globally. The potential roles of CD4(+)T cells in viral control have been extensively studied. However, the contribution of CD4(+)T cells to the host response against Hantaan virus (HTNV) infection remains unclear. Here, based on the T-cell epitopes mapped on HTNV glycoprotein, we studied the effects and characteristics of CD4(+)T-cell responses in determining the outcome of hemorrhagic fever with renal syndrome. A total of 79 novel 15-mer T-cell epitopes on the HTNV glycoprotein were identified, among which 20 peptides were dominant target epitopes. Importantly, we showed the presence of both effective Th1 responses with polyfunctional cytokine secretion and ThGranzyme B(+) cell responses with cytotoxic mediators production against HTNV infection. The HTNV glycoprotein-specific CD4(+)T-cell responses inversely correlated with the plasma HTNV RNA load in patients. Individuals with milder disease outcomes showed broader epitopes targeted and stronger CD4(+)T-cell responses against HTNV glycoproteins compared with more severe patients. The CD4(+)T cells characterized by broader antigenic repertoire, stronger polyfunctional responses, better expansion capacity and highly differentiated effector memory phenotype(CD27-CD28-CCR7-CD45RA-CD127(hi)) would elicit greater defense against HTNV infection and lead to much milder outcome of the disease. The host defense mediated by CD4(+)T cells may through the inducing antiviral condition of the host cells and cytotoxic effect of ThGranzyme B+ cells. Thus, these findings highlight the efforts of CD4(+)T-cell immunity to HTNV control and provide crucial information to better understand the immune defense against HTNV infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, et al. (2013) Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol 11: 539–550. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

