The Drosophila TIPE family member Sigmar interacts with the Ste20-like kinase Misshapen and modulates JNK signaling, cytoskeletal remodeling and autophagy
- PMID: 25836674
- PMCID: PMC4434819
- DOI: 10.1242/bio.20148417
The Drosophila TIPE family member Sigmar interacts with the Ste20-like kinase Misshapen and modulates JNK signaling, cytoskeletal remodeling and autophagy
Abstract
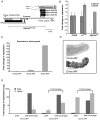
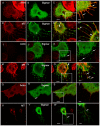
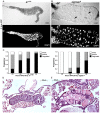
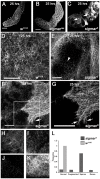
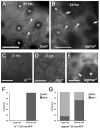
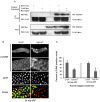
TNFAIP8 and other mammalian TIPE family proteins have attracted increased interest due to their associations with disease-related processes including oncogenic transformation, metastasis, and inflammation. The molecular and cellular functions of TIPE family proteins are still not well understood. Here we report the molecular and genetic characterization of the Drosophila TNFAIP8 homolog, CG4091/sigmar. Previous gene expression studies revealed dynamic expression of sigmar in larval salivary glands prior to histolysis. Here we demonstrate that in sigmar loss-of-function mutants, the salivary glands are morphologically abnormal with defects in the tubulin network and decreased autophagic flux. Sigmar localizes subcellularly to microtubule-containing projections in Drosophila S2 cells, and co-immunoprecipitates with the Ste20-like kinase Misshapen, a regulator of the JNK pathway. Further, the Drosophila TNF ligand Eiger can induce sigmar expression, and sigmar loss-of-function leads to altered localization of pDJNK in salivary glands. Together, these findings link Sigmar to the JNK pathway, cytoskeletal remodeling and autophagy activity during salivary gland development, and provide new insights into TIPE family member function.
Keywords: Autophagy; Cytoskeleton; Eiger; Msn; TIPE; TNFAIP8.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

