Nuclear export of messenger RNA
- PMID: 25836925
- PMCID: PMC4488659
- DOI: 10.3390/genes6020163
Nuclear export of messenger RNA
Abstract
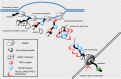
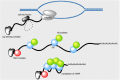
Transport of messenger RNA (mRNA) from the nucleus to the cytoplasm is an essential step of eukaryotic gene expression. In the cell nucleus, a precursor mRNA undergoes a series of processing steps, including capping at the 5' ends, splicing and cleavage/polyadenylation at the 3' ends. During this process, the mRNA associates with a wide variety of proteins, forming a messenger ribonucleoprotein (mRNP) particle. Association with factors involved in nuclear export also occurs during transcription and processing, and thus nuclear export is fully integrated into mRNA maturation. The coupling between mRNA maturation and nuclear export is an important mechanism for providing only fully functional and competent mRNA to the cytoplasmic translational machinery, thereby ensuring accuracy and swiftness of gene expression. This review describes the molecular mechanism of nuclear mRNA export mediated by the principal transport factors, including Tap-p15 and the TREX complex.
Figures

References
-
- Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

