Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking
- PMID: 25837586
- PMCID: PMC4383620
- DOI: 10.1371/journal.pbio.1002097
Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking
Abstract
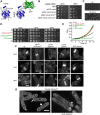
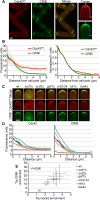
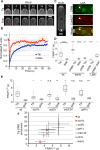
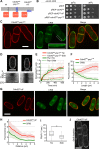
The small Rho-family GTPase Cdc42 is critical for cell polarization and polarizes spontaneously in absence of upstream spatial cues. Spontaneous polarization is thought to require dynamic Cdc42 recycling through Guanine nucleotide Dissociation Inhibitor (GDI)-mediated membrane extraction and vesicle trafficking. Here, we describe a functional fluorescent Cdc42 allele in fission yeast, which demonstrates Cdc42 dynamics and polarization independent of these pathways. Furthermore, an engineered Cdc42 allele targeted to the membrane independently of these recycling pathways by an amphipathic helix is viable and polarizes spontaneously to multiple sites in fission and budding yeasts. We show that Cdc42 is highly mobile at the membrane and accumulates at sites of activity, where it displays slower mobility. By contrast, a near-immobile transmembrane domain-containing Cdc42 allele supports viability and polarized activity, but does not accumulate at sites of activity. We propose that Cdc42 activation, enhanced by positive feedback, leads to its local accumulation by capture of fast-diffusing inactive molecules.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

