Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability
- PMID: 25839327
- PMCID: PMC4385176
- DOI: 10.1016/j.ajhg.2015.02.014
Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability
Abstract
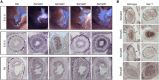
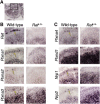
Innervation of the gut is segmentally lost in Hirschsprung disease (HSCR), a consequence of cell-autonomous and non-autonomous defects in enteric neuronal cell differentiation, proliferation, migration, or survival. Rare, high-penetrance coding variants and common, low-penetrance non-coding variants in 13 genes are known to underlie HSCR risk, with the most frequent variants in the ret proto-oncogene (RET). We used a genome-wide association (220 trios) and replication (429 trios) study to reveal a second non-coding variant distal to RET and a non-coding allele on chromosome 7 within the class 3 Semaphorin gene cluster. Analysis in Ret wild-type and Ret-null mice demonstrates specific expression of Sema3a, Sema3c, and Sema3d in the enteric nervous system (ENS). In zebrafish embryos, sema3 knockdowns show reduction of migratory ENS precursors with complete ablation under conjoint ret loss of function. Seven candidate receptors of Sema3 proteins are also expressed within the mouse ENS and their expression is also lost in the ENS of Ret-null embryos. Sequencing of SEMA3A, SEMA3C, and SEMA3D in 254 HSCR-affected subjects followed by in silico protein structure modeling and functional analyses identified five disease-associated alleles with loss-of-function defects in semaphorin dimerization and binding to their cognate neuropilin and plexin receptors. Thus, semaphorin 3C/3D signaling is an evolutionarily conserved regulator of ENS development whose dys-regulation is a cause of enteric aganglionosis.
Copyright © 2015 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Gershon M.D. Harper & Co.; 1998. The Second Brain.
-
- Chakravarti A., Lyonnet S. Hirschsprung disease. In: Scriver C.R., Beaudet A.L., Valle D., Sly W.S., Childs B., Kinzler K., Vogelstein B., editors. The Metabolic and Molecular Bases of Inherited Disease. Eighth Edition. McGraw-Hill; 2001. pp. 6231–6255.
-
- Alves M.M., Sribudiani Y., Brouwer R.W., Amiel J., Antiñolo G., Borrego S., Ceccherini I., Chakravarti A., Fernández R.M., Garcia-Barcelo M.M. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev. Biol. 2013;382:320–329. - PubMed
-
- Emison E.S., McCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., Portnoy M.E., Cutler D.J., Green E.D., Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

