A LCMT1-PME-1 methylation equilibrium controls mitotic spindle size
- PMID: 25839665
- PMCID: PMC4614068
- DOI: 10.1080/15384101.2015.1026487
A LCMT1-PME-1 methylation equilibrium controls mitotic spindle size
Abstract
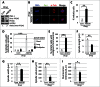
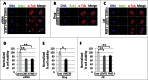
Leucine carboxyl methyltransferase-1 (LCMT1) and protein phosphatase methylesterase-1 (PME-1) are essential enzymes that regulate the methylation of the protein phosphatase 2A catalytic subunit (PP2AC). LCMT1 and PME-1 have been linked to the regulation of cell growth and proliferation, but the underlying mechanisms have remained elusive. We show here an important role for an LCMT1-PME-1 methylation equilibrium in controlling mitotic spindle size. Depletion of LCMT1 or overexpression of PME-1 led to long spindles. In contrast, depletion of PME-1, pharmacological inhibition of PME-1 or overexpression of LCMT1 led to short spindles. Furthermore, perturbation of the LCMT1-PME-1 methylation equilibrium led to mitotic arrest, spindle assembly checkpoint activation, defective cell divisions, induction of apoptosis and reduced cell viability. Thus, we propose that the LCMT1-PME-1 methylation equilibrium is critical for regulating mitotic spindle size and thereby proper cell division.
Keywords: LCMT1; PME-1; cell division; methylation; mitotic spindle.
Figures



References
-
- De Baere I, Derua R, Janssens V, Van Hoof C, Waelkens E, Merlevede W, Goris J. Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry 1999; 38:16539–47; PMID:10600115; http://dx.doi.org/10.1021/bi991646a - DOI - PubMed
-
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell 2009; 33:537–45; PMID:19285938; http://dx.doi.org/10.1016/j.molcel.2009.02.015 - DOI - PubMed
-
- Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V. The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 2013; 280:644–61; PMID:22443683; http://dx.doi.org/10.1111/j.1742-4658.2012.08579.x - DOI - PubMed
-
- Longin S, Zwaenepoel K, Louis JV, Dilworth S, Goris J, Janssens V. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J Biol Chem 2007; 282:26971–80; PMID:17635907; http://dx.doi.org/10.1074/jbc.M704059200 - DOI - PubMed
-
- Ogris E, Du X, Nelson KC, Mak EK, Yu XX, Lane WS, Pallas DC. A protein phosphatase methylesterase (PME-1) is one of several novel proteins stably associating with two inactive mutants of protein phosphatase 2A. J Biol Chem 1999; 274:14382–91; PMID:10318862; http://dx.doi.org/10.1074/jbc.274.20.14382 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
