A Multifunctional Mutagenesis System for Analysis of Gene Function in Zebrafish
- PMID: 25840430
- PMCID: PMC4478556
- DOI: 10.1534/g3.114.015842
A Multifunctional Mutagenesis System for Analysis of Gene Function in Zebrafish
Abstract
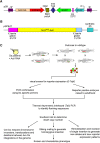
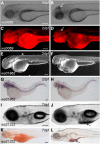
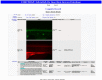
Since the sequencing of the human reference genome, many human disease-related genes have been discovered. However, understanding the functions of all the genes in the genome remains a challenge. The biological activities of these genes are usually investigated in model organisms such as mice and zebrafish. Large-scale mutagenesis screens to generate disruptive mutations are useful for identifying and understanding the activities of genes. Here, we report a multifunctional mutagenesis system in zebrafish using the maize Ds transposon. Integration of the Ds transposable element containing an mCherry reporter for protein trap events and an EGFP reporter for enhancer trap events produced a collection of transgenic lines marking distinct cell and tissue types, and mutagenized genes in the zebrafish genome by trapping and prematurely terminating endogenous protein coding sequences. We obtained 642 zebrafish lines with dynamic reporter gene expression. The characterized fish lines with specific expression patterns will be made available through the European Zebrafish Resource Center (EZRC), and a database of reporter expression is available online (http://fishtrap.warwick.ac.uk/). Our approach complements other efforts using zebrafish to facilitate functional genomic studies in this model of human development and disease.
Keywords: Ac/Ds transposon; functional genomics; gene expression; insertional mutagenesis.
Copyright © 2015 Quach et al.
Figures






Similar articles
-
Generation of an Enhancer-Trapping Vector for Insertional Mutagenesis in Zebrafish.PLoS One. 2015 Oct 5;10(10):e0139612. doi: 10.1371/journal.pone.0139612. eCollection 2015. PLoS One. 2015. PMID: 26436547 Free PMC article.
-
Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like.Development. 2008 Jan;135(1):159-69. doi: 10.1242/dev.009050. Development. 2008. PMID: 18065431
-
zTrap: zebrafish gene trap and enhancer trap database.BMC Dev Biol. 2010 Oct 18;10:105. doi: 10.1186/1471-213X-10-105. BMC Dev Biol. 2010. PMID: 20950494 Free PMC article.
-
[Development of retrovirus-mediated insertional mutagenesis in zebrafish and its application in saturation mutagenesis and gene screening].Yi Chuan Xue Bao. 2004 Jul;31(7):750-7. Yi Chuan Xue Bao. 2004. PMID: 15473329 Review. Chinese.
-
Transposon tools and methods in zebrafish.Dev Dyn. 2005 Oct;234(2):244-54. doi: 10.1002/dvdy.20516. Dev Dyn. 2005. PMID: 16110506 Review.
Cited by
-
Ac/Ds transposition for CRISPR/dCas9-SID4x epigenome modulation in zebrafish.Biol Open. 2023 Jun 15;12(6):bio059995. doi: 10.1242/bio.059995. Epub 2023 Jun 27. Biol Open. 2023. PMID: 37367831 Free PMC article.
-
A native, highly active Tc1/mariner transposon from zebrafish (ZB) offers an efficient genetic manipulation tool for vertebrates.Nucleic Acids Res. 2021 Feb 26;49(4):2126-2140. doi: 10.1093/nar/gkab045. Nucleic Acids Res. 2021. PMID: 33638993 Free PMC article.
-
Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons.Genes (Basel). 2018 Dec 13;9(12):630. doi: 10.3390/genes9120630. Genes (Basel). 2018. PMID: 30551672 Free PMC article.
-
Loss-of-function genetic tools for animal models: cross-species and cross-platform differences.Nat Rev Genet. 2017 Jan;18(1):24-40. doi: 10.1038/nrg.2016.118. Epub 2016 Oct 31. Nat Rev Genet. 2017. PMID: 27795562 Free PMC article. Review.
-
A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish.Nat Protoc. 2016 Dec;11(12):2357-2375. doi: 10.1038/nprot.2016.141. Epub 2016 Oct 27. Nat Protoc. 2016. PMID: 27809318 Free PMC article.
References
-
- Abdelhaleem M., Maltais L., Wain H., 2003. The human DDX and DHX gene families of putative RNA helicases. Genomics 81: 618–622. - PubMed
-
- Allen N. D., Cran D. G., Barton S. C., Hettle S., Reik W., et al. , 1988. Transgenes as probes for active chromosomal domains in mouse development. Nature 333: 852–855. - PubMed
-
- Alli Z., Ackerley C., Chen Y., Al-Saud B., Abdelhaleem M., 2006. Nuclear and mitochondrial localization of the putative RNA helicase DHX32. Exp. Mol. Pathol. 81: 245–248. - PubMed
-
- Boon Ng G. H., Gong Z., 2011. Maize Ac/Ds transposon system leads to highly efficient germline transmission of transgenes in medaka (Oryzias latipes). Biochimie 93: 1858–1864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
