Ionizing radiation induces immediate protein acetylation changes in human cardiac microvascular endothelial cells
- PMID: 25840449
- PMCID: PMC4497387
- DOI: 10.1093/jrr/rrv014
Ionizing radiation induces immediate protein acetylation changes in human cardiac microvascular endothelial cells
Abstract
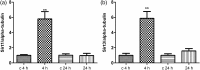
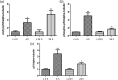
Reversible lysine acetylation is a highly regulated post-translational protein modification that is known to regulate several signaling pathways. However, little is known about the radiation-induced changes in the acetylome. In this study, we analyzed the acute post-translational acetylation changes in primary human cardiac microvascular endothelial cells 4 h after a gamma radiation dose of 2 Gy. The acetylated peptides were enriched using anti-acetyl conjugated agarose beads. A total of 54 proteins were found to be altered in their acetylation status, 23 of which were deacetylated and 31 acetylated. Pathway analyses showed three protein categories particularly affected by radiation-induced changes in the acetylation status: the proteins involved in the translation process, the proteins of stress response, and mitochondrial proteins. The activation of the canonical and non-canonical Wnt signaling pathways affecting actin cytoskeleton signaling and cell cycle progression was predicted. The protein expression levels of two nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases, sirtuin 1 and sirtuin 3, were significantly but transiently upregulated 4 but not 24 h after irradiation. The status of the p53 protein, a target of sirtuin 1, was found to be rapidly stabilized by acetylation after radiation exposure. These findings indicate that post-translational modification of proteins by acetylation and deacetylation is essentially affecting the radiation response of the endothelium.
Keywords: acetylation; endothelial cell; heart; ionizing radiation; proteomics; sirtuins.
© The Author 2015. Published by Oxford University Press on behalf of The Japan Radiation Research Society and Japanese Society for Radiation Oncology.
Figures



References
-
- Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009;325:834–40. - PubMed
-
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 2007;8:284–95. - PubMed
-
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 2007;76:75–100. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 2000;273:793–8. - PubMed
-
- Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free Radic Biol Med 2013;56:133–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

