The changing face of asthma and its relation with microbes
- PMID: 25840766
- PMCID: PMC4710578
- DOI: 10.1016/j.tim.2015.03.005
The changing face of asthma and its relation with microbes
Abstract
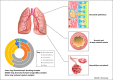
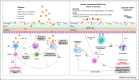
During the past 50 years, the prevalence of asthma has increased and this has coincided with our changing relation with microorganisms. Asthma is a complex disease associated with local tissue inflammation of the airway that is determined by environmental, immunological, and host genetic factors. In a subgroup of sufferers, respiratory infections are associated with the development of chronic disease and more frequent inflammatory exacerbations. Recent studies suggest that these infections are polymicrobial in nature. Furthermore, there is increasing evidence that the recently discovered asthma airway microbiota may play a critical role in pathophysiological processes associated with the disease. Here, we discuss the current data regarding a possible role for infection in chronic asthma with a particular focus on the role bacteria may play. We discuss recent advances that are beginning to elucidate the complex relations between the microbiota and the immune response in asthma patients. We also highlight the clinical implications of these recent findings in regards to the development of novel therapeutic strategies.
Keywords: airway; allergens; asthma; infection; inflammation; microbiota.
Crown Copyright © 2015. Published by Elsevier Ltd. All rights reserved.
Figures
References
-
- Kopf M. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 2015;16:36–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

