Differential Metabolism of Exopolysaccharides from Probiotic Lactobacilli by the Human Gut Symbiont Bacteroides thetaiotaomicron
- PMID: 25841008
- PMCID: PMC4524142
- DOI: 10.1128/AEM.00149-15
Differential Metabolism of Exopolysaccharides from Probiotic Lactobacilli by the Human Gut Symbiont Bacteroides thetaiotaomicron
Abstract
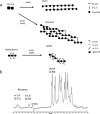
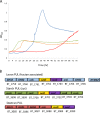
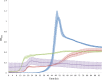
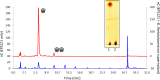
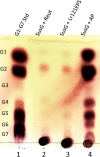
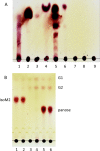
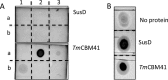
Probiotic microorganisms are ingested as food or supplements and impart positive health benefits to consumers. Previous studies have indicated that probiotics transiently reside in the gastrointestinal tract and, in addition to modulating commensal species diversity, increase the expression of genes for carbohydrate metabolism in resident commensal bacterial species. In this study, it is demonstrated that the human gut commensal species Bacteroides thetaiotaomicron efficiently metabolizes fructan exopolysaccharide (EPS) synthesized by probiotic Lactobacillus reuteri strain 121 while only partially degrading reuteran and isomalto/malto-polysaccharide (IMMP) α-glucan EPS polymers. B. thetaiotaomicron metabolized these EPS molecules via the activation of enzymes and transport systems encoded by dedicated polysaccharide utilization loci specific for β-fructans and α-glucans. Reduced metabolism of reuteran and IMMP α-glucan EPS molecules may be due to reduced substrate binding by components of the starch utilization system (sus). This study reveals that microbial EPS substrates activate genes for carbohydrate metabolism in B. thetaiotaomicron and suggests that microbially derived carbohydrates provide a carbohydrate-rich reservoir for B. thetaiotaomicron nutrient acquisition in the gastrointestinal tract.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis.mBio. 2014 Sep 9;5(5):e01441-14. doi: 10.1128/mBio.01441-14. mBio. 2014. PMID: 25205092 Free PMC article.
-
Bacteroides thetaiotaomicron metabolic activity decreases with polysaccharide molecular weight.mBio. 2024 Mar 13;15(3):e0259923. doi: 10.1128/mbio.02599-23. Epub 2024 Feb 20. mBio. 2024. PMID: 38376161 Free PMC article.
-
Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts.mBio. 2014 Nov 11;5(6):e02172. doi: 10.1128/mBio.02172-14. mBio. 2014. PMID: 25389179 Free PMC article.
-
Navigating the Gut Buffet: Control of Polysaccharide Utilization in Bacteroides spp.Trends Microbiol. 2017 Dec;25(12):1005-1015. doi: 10.1016/j.tim.2017.06.009. Epub 2017 Jul 18. Trends Microbiol. 2017. PMID: 28733133 Review.
-
The Sus operon: a model system for starch uptake by the human gut Bacteroidetes.Cell Mol Life Sci. 2016 Jul;73(14):2603-17. doi: 10.1007/s00018-016-2242-x. Epub 2016 May 2. Cell Mol Life Sci. 2016. PMID: 27137179 Free PMC article. Review.
Cited by
-
Carbohydrate esterases involved in deacetylation of food components by the human gut microbiota.Essays Biochem. 2023 Apr 18;67(3):443-454. doi: 10.1042/EBC20220161. Essays Biochem. 2023. PMID: 36912209 Free PMC article.
-
Characterization of the bacterial microbiota composition and evolution at different intestinal tract in wild pigs (Sus scrofa ussuricus).PeerJ. 2020 May 26;8:e9124. doi: 10.7717/peerj.9124. eCollection 2020. PeerJ. 2020. PMID: 32518722 Free PMC article.
-
Intestinal microbiota and chronic constipation.Springerplus. 2016 Jul 19;5(1):1130. doi: 10.1186/s40064-016-2821-1. eCollection 2016. Springerplus. 2016. PMID: 27478747 Free PMC article. Review.
-
Construction and characterization of a genome-scale ordered mutant collection of Bacteroides thetaiotaomicron.BMC Biol. 2022 Dec 17;20(1):285. doi: 10.1186/s12915-022-01481-2. BMC Biol. 2022. PMID: 36527020 Free PMC article.
-
Characterization of a glycoside hydrolase family 78 α-l-rhamnosidase from Bacteroides thetaiotaomicron VPI-5482 and identification of functional residues.3 Biotech. 2018 Feb;8(2):120. doi: 10.1007/s13205-018-1139-9. Epub 2018 Feb 8. 3 Biotech. 2018. PMID: 29430381 Free PMC article.
References
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi:10.1038/nature08821. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

