Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY)
- PMID: 25844078
- PMCID: PMC4379436
- DOI: 10.1107/S160057671500254X
Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY)
Abstract
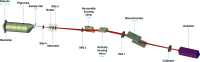
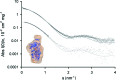
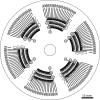
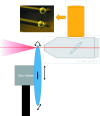
A high-brilliance synchrotron P12 beamline of the EMBL located at the PETRA III storage ring (DESY, Hamburg) is dedicated to biological small-angle X-ray scattering (SAXS) and has been designed and optimized for scattering experiments on macromolecular solutions. Scatterless slits reduce the parasitic scattering, a custom-designed miniature active beamstop ensures accurate data normalization and the photon-counting PILATUS 2M detector enables the background-free detection of weak scattering signals. The high flux and small beam size allow for rapid experiments with exposure time down to 30-50 ms covering the resolution range from about 300 to 0.5 nm. P12 possesses a versatile and flexible sample environment system that caters for the diverse experimental needs required to study macromolecular solutions. These include an in-vacuum capillary mode for standard batch sample analyses with robotic sample delivery and for continuous-flow in-line sample purification and characterization, as well as an in-air capillary time-resolved stopped-flow setup. A novel microfluidic centrifugal mixing device (SAXS disc) is developed for a high-throughput screening mode using sub-microlitre sample volumes. Automation is a key feature of P12; it is controlled by a beamline meta server, which coordinates and schedules experiments from either standard or nonstandard operational setups. The integrated SASFLOW pipeline automatically checks for consistency, and processes and analyses the data, providing near real-time assessments of overall parameters and the generation of low-resolution models within minutes of data collection. These advances, combined with a remote access option, allow for rapid high-throughput analysis, as well as time-resolved and screening experiments for novice and expert biological SAXS users.
Keywords: automated hardware and software systems; biological small-angle X-ray scattering; high-brilliance P12 synchrotron beamline.
Figures



References
-
- Bartkiewicz, P. & Duval, P. (2007). Meas. Sci. Technol. 18, 2379–2386.
-
- Blanchet, C. E. & Svergun, D. I. (2013). Annu. Rev. Phys. Chem. 64, 37–54. - PubMed
-
- Blanchet, C. E., Zozulya, A. V., Kikhney, A. G., Franke, D., Konarev, P. V., Shang, W., Klaering, R., Robrahn, B., Hermes, C., Cipriani, F., Svergun, D. I. & Roessle, M. (2012). J. Appl. Cryst. 45, 489–495.
-
- Callender, R. & Dyer, R. B. (2006). Chem. Rev. 106, 3031–3042. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
