Site-specific DNA Inversion by Serine Recombinases
- PMID: 25844275
- PMCID: PMC4384473
- DOI: 10.1128/microbiolspec.MDNA3-0047-2014
Site-specific DNA Inversion by Serine Recombinases
Abstract
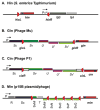
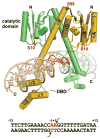
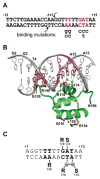
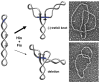
Reversible site-specific DNA inversion reactions are widely distributed in bacteria and their viruses. They control a range of biological reactions that most often involve alterations of molecules on the surface of cells or phage. These programmed DNA rearrangements usually occur at a low frequency, thereby preadapting a small subset of the population to a change in environmental conditions, or in the case of phages, an expanded host range. A dedicated recombinase, sometimes with the aid of additional regulatory or DNA architectural proteins, catalyzes the inversion of DNA. RecA or other components of the general recombination-repair machinery are not involved. This chapter discusses site-specific DNA inversion reactions mediated by the serine recombinase family of enzymes and focuses on the extensively studied serine DNA invertases that are stringently controlled by the Fis-bound enhancer regulatory system. The first section summarizes biological features and general properties of inversion reactions by the Fis/enhancer-dependent serine invertases and the recently described serine DNA invertases in Bacteroides. Mechanistic studies of reactions catalyzed by the Hin and Gin invertases are then discussed in more depth, particularly with regards to recent advances in our understanding of the function of the Fis/enhancer regulatory system, the assembly of the active recombination complex (invertasome) containing the Fis/enhancer, and the process of DNA strand exchange by rotation of synapsed subunit pairs within the invertasome. The role of DNA topological forces that function in concert with the Fis/enhancer controlling element in specifying the overwhelming bias for DNA inversion over deletion and intermolecular recombination is emphasized.
Figures






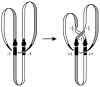
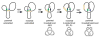




Similar articles
-
Site-specific DNA Inversion by Serine Recombinases.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0047-2014. doi: 10.1128/microbiolspec.MDNA3-0047-2014. Microbiol Spectr. 2015. PMID: 26104558 Review.
-
The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion.Curr Biol. 1996 Feb 1;6(2):163-77. doi: 10.1016/s0960-9822(02)00449-9. Curr Biol. 1996. PMID: 8673463
-
Multiple interfaces between a serine recombinase and an enhancer control site-specific DNA inversion.Elife. 2013 Oct 22;2:e01211. doi: 10.7554/eLife.01211. Elife. 2013. PMID: 24151546 Free PMC article.
-
Communication between Hin recombinase and Fis regulatory subunits during coordinate activation of Hin-catalyzed site-specific DNA inversion.Genes Dev. 1998 Sep 1;12(17):2803-16. doi: 10.1101/gad.12.17.2803. Genes Dev. 1998. PMID: 9732277 Free PMC article.
-
The Fis protein: it's not just for DNA inversion anymore.Mol Microbiol. 1992 Nov;6(22):3257-65. doi: 10.1111/j.1365-2958.1992.tb02193.x. Mol Microbiol. 1992. PMID: 1484481 Review.
Cited by
-
Comparative genome and phenotypic analysis of three Clostridioides difficile strains isolated from a single patient provide insight into multiple infection of C. difficile.BMC Genomics. 2018 Jan 2;19(1):1. doi: 10.1186/s12864-017-4368-0. BMC Genomics. 2018. PMID: 29291715 Free PMC article.
-
Controlled rotation mechanism of DNA strand exchange by the Hin serine recombinase.Sci Rep. 2016 Apr 1;6:23697. doi: 10.1038/srep23697. Sci Rep. 2016. PMID: 27032966 Free PMC article.
-
Remnants of horizontal transfers of Wolbachia genes in a Wolbachia-free woodwasp.BMC Ecol Evol. 2022 Mar 26;22(1):36. doi: 10.1186/s12862-022-01995-x. BMC Ecol Evol. 2022. PMID: 35346038 Free PMC article.
-
Creation of a eukaryotic multiplexed site-specific inversion system and its application for metabolic engineering.Nat Commun. 2025 Feb 24;16(1):1918. doi: 10.1038/s41467-025-57206-w. Nat Commun. 2025. PMID: 39994248 Free PMC article.
-
Recruitment of Mobile Genetic Elements for Diverse Cellular Functions in Prokaryotes.Front Mol Biosci. 2022 Mar 24;9:821197. doi: 10.3389/fmolb.2022.821197. eCollection 2022. Front Mol Biosci. 2022. PMID: 35402511 Free PMC article. Review.
References
-
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. - PubMed
-
- Johnson RC. Bacterial site-specific DNA inversion systems. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, D.C: 2002. pp. 230–271.
-
- Rice PA. Title to be added. In: Craig NL, editor. Mobile DNA III. ASM Press; Washington, D.C: 2014. in press.
-
- Stark WM. The serine recombinases. In: Craig NL, editor. Mobile DNA III. ASM Press; Washington, D.C: 2014. in press.
-
- Andrewes FW. Studies in group-agglutination - The Salmonella group and its antigenic structure. J Pathology and Bacteriology. 1922;25:505–521.
Grants and funding
LinkOut - more resources
Full Text Sources

