PRMT1 Is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-small Cell Lung Cancer
- PMID: 25847239
- PMCID: PMC4505594
- DOI: 10.1074/jbc.M114.636050
PRMT1 Is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-small Cell Lung Cancer
Abstract
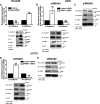
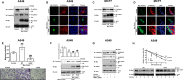
Protein arginine methyl transferase 1 (PRMT1) was shown to be up-regulated in cancers and important for cancer cell proliferation. However, the role of PRMT1 in lung cancer progression and metastasis remains incompletely understood. In the present study, we show that PRMT1 is an important regulator of epithelial-mesenchymal transition (EMT), cancer cell migration, and invasion, which are essential processes during cancer progression, and metastasis. Additionally, we have identified Twist1, a basic helix-loop-helix transcription factor and a well-known E-cadherin repressor, as a novel PRMT1 substrate. Taken together, we show that PRMT1 is a novel regulator of EMT and arginine 34 (Arg-34) methylation of Twist1 as a unique "methyl arginine mark" for active E-cadherin repression. Therefore, targeting PRMT1-mediated Twist1 methylation might represent a novel strategy for developing new anti-invasive/anti-metastatic drugs. Moreover, methylated Twist1 (Arg-34), as such, could also emerge as a potential important biomarker for lung cancer.
Keywords: N-cadherin; Prmt1; Twist1; cadherin-1 (CDH1) (epithelial cadherin) (E-cadherin); epithelial-mesenchymal transition (EMT); metastasis; protein methylation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Yang Y., Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nature Reviews. Cancer 13, 37–50 - PubMed
-
- Cha B., Jho E. H. (2012) Protein arginine methyltransferases (PRMTs) as therapeutic targets. Exp. Opin. Therap. Targets 16, 651–664 - PubMed
-
- Paik W. K., Kim S. (1968) Protein methylase I. Purification and properties of the enzyme. J. Biol. Chem. 243, 2108–2114 - PubMed
-
- Lin W. J., Gary J. D., Yang M. C., Clarke S., Herschman H. R. (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271, 15034–15044 - PubMed
-
- Boffa L. C., Karn J., Vidali G., Allfrey V. G. (1977) Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem. Biophys. Res. Commun. 74, 969–976 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

