Architecture of the cerebral cortical association connectome underlying cognition
- PMID: 25848037
- PMCID: PMC4413280
- DOI: 10.1073/pnas.1504394112
Architecture of the cerebral cortical association connectome underlying cognition
Abstract
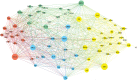
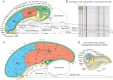
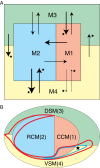
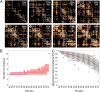
Cognition presumably emerges from neural activity in the network of association connections between cortical regions that is modulated by inputs from sensory and state systems and directs voluntary behavior by outputs to the motor system. To reveal global architectural features of the cortical association connectome, network analysis was performed on >16,000 reports of histologically defined axonal connections between cortical regions in rat. The network analysis reveals an organization into four asymmetrically interconnected modules involving the entire cortex in a topographic and topologic core-shell arrangement. There is also a topographically continuous U-shaped band of cortical areas that are highly connected with each other as well as with the rest of the cortex extending through all four modules, with the temporal pole of this band (entorhinal area) having the most cortical association connections of all. These results provide a starting point for compiling a mammalian nervous system connectome that could ultimately reveal novel correlations between genome-wide association studies and connectome-wide association studies, leading to new insights into the cellular architecture supporting cognition.
Keywords: cerebral cortex; connectomics; mammal; network analysis; neural connections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kandel ER, Hudspeth AJ. The brain and behavior. In: Kandel ER, Schwartz JH, Jessell TM, Siegelbaum SA, Hudspeth AJ, editors. Principles of Neural Science. 5th Ed. McGraw-Hill; New York: 2013. pp. 5–20.
-
- Swanson LW. Brain Architecture: Understanding the Basic Plan. 2nd Ed Oxford Univ Press; Oxford: 2012.
-
- Catani M, Mesulam M. What is a disconnection syndrome? Cortex. 2008;44(8):911–913. - PubMed
-
- Nieuwenhuys R, Voogd J, van Huijzen C. The Human Central Nervous System. 4th Ed Springer; Berlin: 2008.
-
- Bota M, Dong H-W, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6(8):795–799. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

