FtsZ filament capping by MciZ, a developmental regulator of bacterial division
- PMID: 25848052
- PMCID: PMC4418908
- DOI: 10.1073/pnas.1414242112
FtsZ filament capping by MciZ, a developmental regulator of bacterial division
Abstract
Cytoskeletal structures are dynamically remodeled with the aid of regulatory proteins. FtsZ (filamentation temperature-sensitive Z) is the bacterial homolog of tubulin that polymerizes into rings localized to cell-division sites, and the constriction of these rings drives cytokinesis. Here we investigate the mechanism by which the Bacillus subtilis cell-division inhibitor, MciZ (mother cell inhibitor of FtsZ), blocks assembly of FtsZ. The X-ray crystal structure reveals that MciZ binds to the C-terminal polymerization interface of FtsZ, the equivalent of the minus end of tubulin. Using in vivo and in vitro assays and microscopy, we show that MciZ, at substoichiometric levels to FtsZ, causes shortening of protofilaments and blocks the assembly of higher-order FtsZ structures. The findings demonstrate an unanticipated capping-based regulatory mechanism for FtsZ.
Keywords: FtsZ; bacterial cytoskeleton; cell division; cytokinesis; filament capping.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
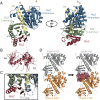
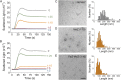

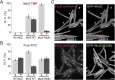
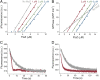

References
-
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354(6349):161–164. - PubMed
-
- Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5(6):451–458. - PubMed
-
- Adams DW, Errington J. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7(9):642–653. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

