Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function
- PMID: 25848867
- PMCID: PMC4406853
- DOI: 10.1038/ni.3146
Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function
Abstract
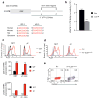
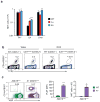
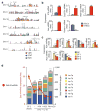
Lethal-7 (let-7) microRNAs (miRNAs) are the most abundant miRNAs in the genome, but their role in developing thymocytes is unclear. We found that let-7 miRNAs targeted Zbtb16 mRNA, which encodes the lineage-specific transcription factor PLZF, to post-transcriptionally regulate PLZF expression and thereby the effector functions of natural killer T cells (NKT cells). Dynamic upregulation of let-7 miRNAs during the development of NKT thymocytes downregulated PLZF expression and directed their terminal differentiation into interferon-γ (IFN-γ)-producing NKT1 cells. Without upregulation of let-7 miRNAs, NKT thymocytes maintained high PLZF expression and terminally differentiated into interleukin 4 (IL-4)-producing NKT2 cells or IL-17-producing NKT17 cells. Upregulation of let-7 miRNAs in developing NKT thymocytes was signaled by IL-15, vitamin D and retinoic acid. Such targeting of a lineage-specific transcription factor by miRNA represents a previously unknown level of developmental regulation in the thymus.
Figures




References
-
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. - PubMed
-
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. - PubMed
-
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. - PubMed
-
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

