Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi
- PMID: 25848876
- PMCID: PMC4579475
- DOI: 10.1038/ismej.2015.35
Growth phase-specific evolutionary benefits of natural transformation in Acinetobacter baylyi
Abstract
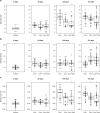
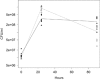
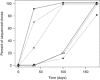
Natural transformation in bacteria facilitates the uptake and genomic integration of exogenous DNA. This allows horizontal exchange of adaptive traits not easily achieved by point mutations, and has a major role in the acquisition of adaptive traits exemplified by antibiotic resistance determinants and vaccination escape. Mechanisms of DNA uptake and genomic integration are well described for several naturally transformable bacterial species; however, the selective forces responsible for its evolution and maintenance are still controversial. In this study we evolved transformation-proficient and -deficient Acinetobacter baylyi for 175 days in serial transfer cultures where stress was included. We found that natural transformation-proficient populations adapted better to active growth and early stationary phase. This advantage was offset by the reduced performance in the late stationary/death phase. We demonstrate fitness trade-offs between adaptation to active growth and survival in stationary/death phase caused by antagonistic pleiotropy. The presented data suggest that the widely held assumption that recombination speeds up adaptation by rapid accumulation of multiple adaptive mutations in the same genetic background is not sufficient to fully account for the maintenance of natural transformation in bacteria.
Figures


References
-
- Averhoff B, Graf I. (2008). The Natural Transformation System of Acinetobacter baylyi ADP1: A Unique DNA Transport Machinery. In: Gerischer U (ed). Acinetobacter—Molecular Biology. Caister Academic Press: Norfolk, UK, pp 119–139.
-
- Baltrus D. (2013). Exploring the costs of horizontal gene transfer. Trends Ecol Evol 28: 489–495. - PubMed
-
- Baltrus D, Guillemin K, Phillips P. (2007). Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62: 10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

