Parallel action of AtDRB2 and RdDM in the control of transposable element expression
- PMID: 25849103
- PMCID: PMC4351826
- DOI: 10.1186/s12870-015-0455-z
Parallel action of AtDRB2 and RdDM in the control of transposable element expression
Abstract
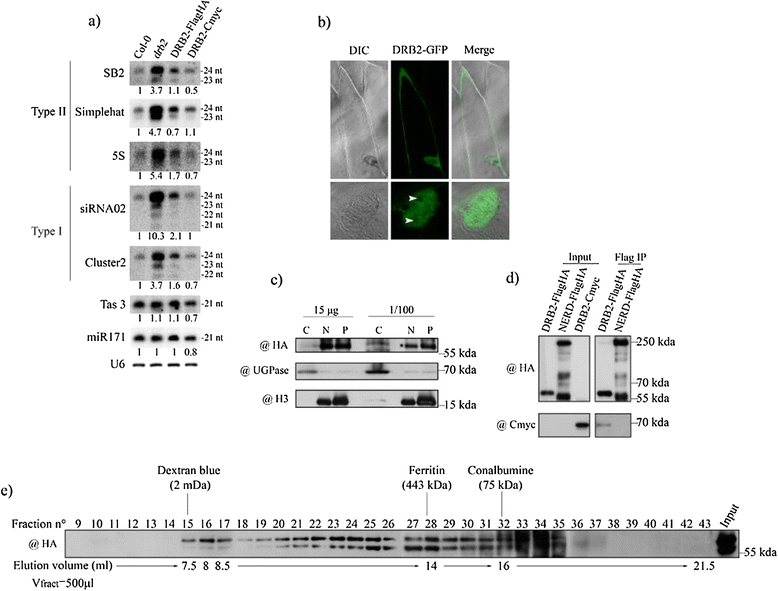
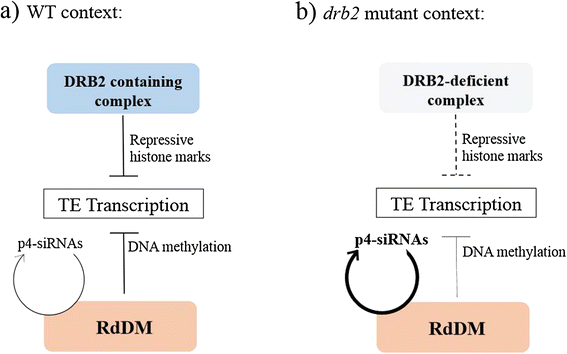
Background: In plants and animals, a large number of double-stranded RNA binding proteins (DRBs) have been shown to act as non-catalytic cofactors of DICERs and to participate in the biogenesis of small RNAs involved in RNA silencing. We have previously shown that the loss of Arabidopsis thaliana's DRB2 protein results in a significant increase in the population of RNA polymerase IV (p4) dependent siRNAs, which are involved in the RNA-directed DNA methylation (RdDM) process.
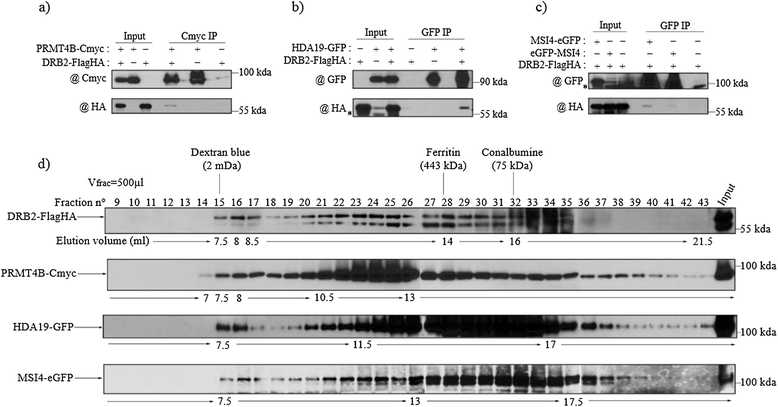
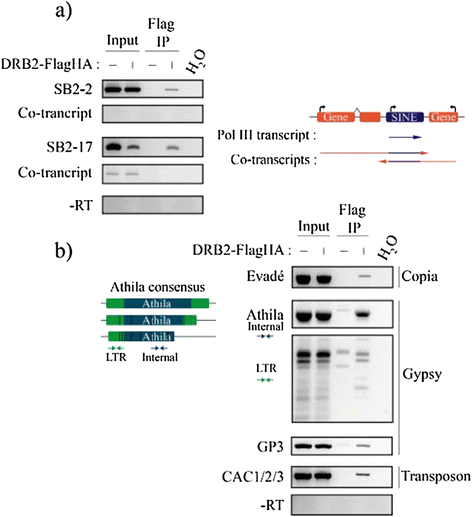
Results: Surprisingly, despite this observation, we show in this work that DRB2 is part of a high molecular weight complex that does not involve RdDM actors but several chromatin regulator proteins, such as MSI4, PRMT4B and HDA19. We show that DRB2 can bind transposable element (TE) transcripts in vivo but that drb2 mutants do not have a significant variation in TE DNA methylation.
Conclusion: We propose that DRB2 is part of a repressive epigenetic regulator complex involved in a negative feedback loop, adjusting epigenetic state to transcription level at TE loci, in parallel of the RdDM pathway. Loss of DRB2 would mainly result in an increased production of TE transcripts, readily converted in p4-siRNAs by the RdDM machinery.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

