Domain movements during CCA-addition: a new function for motif C in the catalytic core of the human tRNA nucleotidyltransferases
- PMID: 25849199
- PMCID: PMC4615804
- DOI: 10.1080/15476286.2015.1018502
Domain movements during CCA-addition: a new function for motif C in the catalytic core of the human tRNA nucleotidyltransferases
Abstract
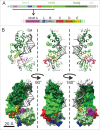
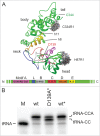
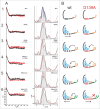
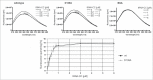
CCA-adding enzymes are highly specific RNA polymerases that synthesize and maintain the sequence CCA at the tRNA 3'-end. This nucleotide triplet is a prerequisite for tRNAs to be aminoacylated and to participate in protein biosynthesis. During CCA-addition, a set of highly conserved motifs in the catalytic core of these enzymes is responsible for accurate sequential nucleotide incorporation. In the nucleotide binding pocket, three amino acid residues form Watson-Crick-like base pairs to the incoming CTP and ATP. A reorientation of these templating amino acids switches the enzyme's specificity from CTP to ATP recognition. However, the mechanism underlying this essential structural rearrangement is not understood. Here, we show that motif C, whose actual function has not been identified yet, contributes to the switch in nucleotide specificity during polymerization. Biochemical characterization as well as EPR spectroscopy measurements of the human enzyme reveal that mutating the highly conserved amino acid position D139 in this motif interferes with AMP incorporation and affects interdomain movements in the enzyme. We propose a model of action, where motif C forms a flexible spring element modulating the relative orientation of the enzyme's head and body domains to accommodate the growing 3'-end of the tRNA. Furthermore, these conformational transitions initiate the rearranging of the templating amino acids to switch the specificity of the nucleotide binding pocket from CTP to ATP during CCA-synthesis.
Keywords: CCA-adding enzyme; CCA-addition; DEER; EPR spectroscopy; tRNA; tRNA maturation; tRNA nucleotidyltransferase.
Figures

Similar articles
-
A single catalytically active subunit in the multimeric Sulfolobus shibatae CCA-adding enzyme can carry out all three steps of CCA addition.J Biol Chem. 2004 Sep 17;279(38):40130-6. doi: 10.1074/jbc.M405518200. Epub 2004 Jul 19. J Biol Chem. 2004. PMID: 15265870
-
Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase.Structure. 2014 Feb 4;22(2):315-25. doi: 10.1016/j.str.2013.12.002. Epub 2014 Jan 2. Structure. 2014. PMID: 24389024
-
Mechanism of 3'-Matured tRNA Discrimination from 3'-Immature tRNA by Class-II CCA-Adding Enzyme.Structure. 2016 Jun 7;24(6):918-25. doi: 10.1016/j.str.2016.03.022. Epub 2016 Apr 28. Structure. 2016. PMID: 27133023
-
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.Cell Mol Life Sci. 2010 May;67(9):1447-63. doi: 10.1007/s00018-010-0271-4. Epub 2010 Feb 14. Cell Mol Life Sci. 2010. PMID: 20155482 Free PMC article. Review.
-
The CCA-adding enzyme: A central scrutinizer in tRNA quality control.Bioessays. 2015 Sep;37(9):975-82. doi: 10.1002/bies.201500043. Epub 2015 Jul 14. Bioessays. 2015. PMID: 26172425 Review.
Cited by
-
Unusual Occurrence of Two Bona-Fide CCA-Adding Enzymes in Dictyostelium discoideum.Int J Mol Sci. 2020 Jul 23;21(15):5210. doi: 10.3390/ijms21155210. Int J Mol Sci. 2020. PMID: 32717856 Free PMC article.
-
CCA-Addition Gone Wild: Unusual Occurrence and Phylogeny of Four Different tRNA Nucleotidyltransferases in Acanthamoeba castellanii.Mol Biol Evol. 2021 Mar 9;38(3):1006-1017. doi: 10.1093/molbev/msaa270. Mol Biol Evol. 2021. PMID: 33095240 Free PMC article.
-
Divergent Evolution of Eukaryotic CC- and A-Adding Enzymes.Int J Mol Sci. 2020 Jan 10;21(2):462. doi: 10.3390/ijms21020462. Int J Mol Sci. 2020. PMID: 31936900 Free PMC article.
-
Adaptation of the Romanomermis culicivorax CCA-Adding Enzyme to Miniaturized Armless tRNA Substrates.Int J Mol Sci. 2020 Nov 28;21(23):9047. doi: 10.3390/ijms21239047. Int J Mol Sci. 2020. PMID: 33260740 Free PMC article.
-
Two complementing in vivo selection systems based on CCA-trimming exonucleases as a tool to monitor, select and evaluate enzymatic features of tRNA nucleotidyltransferases.RNA Biol. 2025 Dec;22(1):1-14. doi: 10.1080/15476286.2025.2453963. Epub 2025 Jan 29. RNA Biol. 2025. PMID: 39831457 Free PMC article.
References
-
- Deutscher MP. tRNA Nucleotidyltransferase. Boyer PD ed Nucl Acid Part B Acad Press 1982:183-215
-
- Sprinzl M, Cramer F. The-C-C-A end of tRNA and its role in protein biosynthesis. Prog Nucl Acid Res Mol Biol 1979; 22:1-69; PMID:392600; http://dx.doi.org/10.1016/S0079-6603(08)60798-9 - DOI - PubMed
-
- Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trend Biochem Sci 1995; 20:345-7; PMID:7482698 - PubMed
-
- Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA 2007; 13:1834-49; PMID:17872511; http://dx.doi.org/10.1261/rna.652807 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
