Prevalence study and genetic typing of bovine viral diarrhea virus (BVDV) in four bovine species in China
- PMID: 25849315
- PMCID: PMC4388703
- DOI: 10.1371/journal.pone.0121718
Prevalence study and genetic typing of bovine viral diarrhea virus (BVDV) in four bovine species in China
Erratum in
-
Correction: Prevalence Study and Genetic Typing of Bovine Viral Diarrhea Virus (BVDV) in Four Bovine Species in China.PLoS One. 2015 Jul 31;10(7):e0134777. doi: 10.1371/journal.pone.0134777. eCollection 2015. PLoS One. 2015. PMID: 26230393 Free PMC article. No abstract available.
Abstract
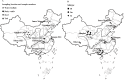
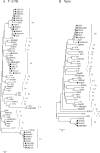
To determine the nationwide status of persistent BVDV infection in different bovine species in China and compare different test methods, a total of 1379 serum samples from clinical healthy dairy cattle, beef cattle, yaks (Bos grunniens), and water buffalo (Bubalus bubalis) were collected in eight provinces of China from 2010 to 2013. The samples were analyzed using commercial antibody (Ab) and antigen (Ag) detection kits, and RT-PCR based on the 5'-UTR and Npro gene sequencing. Results showed that the overall positive rates for BVDV Ab, Ag and RT-PCR detection were 58.09% (801/1379), 1.39% (14/1010), and 22.64% (146/645), respectively, while the individual positive rates varied among regions, species, and farms. The average Ab-positive rates for dairy cattle, beef cattle, yaks, and water buffalo were 89.49% (298/333), 63.27% (248/392), 45.38% (236/520), and 14.18% (19/134), respectively, while the Ag-positive rates were 0.00% (0/116), 0.77% (3/392), 0.82% (3/368), and 5.97% (8/134), respectively, and the nucleic acid-positive rates detected by RT-PCR were 32.06% (42/131), 13.00% (26/200), 28.89% (52/180), and 19.40% (26/134), respectively. In addition, the RT-PCR products were sequenced and 124 5'-UTR sequences were obtained. Phylogenetic analysis of the 5'-UTR sequences indicated that all of the 124 BVDV-positive samples were BVDV-1 and subtyped into either BVDV-1b (33.06%), BVDV-1m (49.19%), or a new cluster, designated as BVDV-1u (17.74%). Phylogenetic analysis based on Npro sequences confirmed this novel subtype. In conclusion, this study revealed the prevalence of BVDV-1 in bovine species in China and the dominant subtypes. The high proportion of bovines with detectable viral nucleic acids in the sera, even in the presence of high Ab levels, revealed a serious threat to bovine health.
Conflict of interest statement
Figures
Similar articles
-
Prevalence and genetic diversity of bovine viral diarrhea virus in dairy herds of China.Vet Microbiol. 2020 Mar;242:108565. doi: 10.1016/j.vetmic.2019.108565. Epub 2019 Dec 27. Vet Microbiol. 2020. PMID: 32122580
-
Molecular analyses detect natural coinfection of water buffaloes (Bubalus bubalis) with bovine viral diarrhea viruses (BVDV) in serologically negative animals.Rev Argent Microbiol. 2015 Apr-Jun;47(2):148-51. doi: 10.1016/j.ram.2015.03.001. Epub 2015 May 8. Rev Argent Microbiol. 2015. PMID: 25962538
-
Genotyping and phylogenetic analysis of bovine viral diarrhea virus isolates from BVDV infected alpacas in North America.Vet Microbiol. 2009 May 12;136(3-4):209-16. doi: 10.1016/j.vetmic.2008.10.029. Epub 2008 Nov 5. Vet Microbiol. 2009. PMID: 19059738
-
A systematic review and meta-analysis of the epidemiology of bovine viral diarrhea virus (BVDV) infection in dairy cattle in China.Acta Trop. 2019 Feb;190:296-303. doi: 10.1016/j.actatropica.2018.08.031. Epub 2018 Aug 27. Acta Trop. 2019. PMID: 30165071
-
Prevalence of bovine viral diarrhea virus (BVDV) in yaks between 1987 and 2019 in mainland China: A systematic review and meta-analysis.Microb Pathog. 2020 Jul;144:104185. doi: 10.1016/j.micpath.2020.104185. Epub 2020 Apr 6. Microb Pathog. 2020. PMID: 32272215
Cited by
-
Detection of emerging HoBi-like Pestivirus (BVD-3) during an epidemiological investigation of bovine viral diarrhea virus in Xinjiang: a first-of-its-kind report.Front Microbiol. 2023 Jul 4;14:1222292. doi: 10.3389/fmicb.2023.1222292. eCollection 2023. Front Microbiol. 2023. PMID: 37492265 Free PMC article.
-
Molecular Characteristics of Bovine Viral Diarrhea Virus Strains Isolated from Persistently Infected Cattle.Vet Sci. 2023 Jun 25;10(7):413. doi: 10.3390/vetsci10070413. Vet Sci. 2023. PMID: 37505819 Free PMC article.
-
Identification and genotyping of a new subtype of bovine viral diarrhea virus 1 isolated from cattle with diarrhea.Arch Virol. 2021 Apr;166(4):1259-1262. doi: 10.1007/s00705-021-04990-7. Epub 2021 Feb 13. Arch Virol. 2021. PMID: 33582856 Free PMC article.
-
Survey and Associated Risk Factors for the Presence of Ruminant Pestiviruses in Domestic Ovine and Caprine Populations from Kazakhstan.Viruses. 2025 May 6;17(5):676. doi: 10.3390/v17050676. Viruses. 2025. PMID: 40431688 Free PMC article.
-
Isolation of BVDV-1a, 1m, and 1v strains from diarrheal calf in china and identification of its genome sequence and cattle virulence.Front Vet Sci. 2022 Nov 16;9:1008107. doi: 10.3389/fvets.2022.1008107. eCollection 2022. Front Vet Sci. 2022. PMID: 36467650 Free PMC article.
References
-
- Lee KM, Gillespie JH (1957) Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res 18: 952–953. - PubMed
-
- Baker JC (1995) The clinical manifestations of bovine viral diarrhea infection. Vet Clin North Am Food Anim Pract 11: 425–445. - PubMed
-
- Brownlie J, Clarke MC, Howard CJ (1984) Experimental production of fatal mucosal disease in cattle. Vet Rec 114: 535–536. - PubMed
-
- Coria MF, McClurkin AW (1978) Specific immune tolerance in an apparently healthy bull persistently infected with bovine viral diarrhea virus. J Am Vet Med Assoc 172: 449–451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

