The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta)
- PMID: 25849557
- PMCID: PMC4388391
- DOI: 10.1371/journal.pone.0121020
The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta)
Abstract
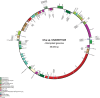
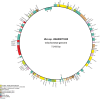
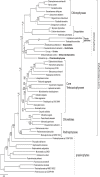
Sequencing mitochondrial and chloroplast genomes has become an integral part in understanding the genomic machinery and the phylogenetic histories of green algae. Previously, only three chloroplast genomes (Oltmannsiellopsis viridis, Pseudendoclonium akinetum, and Bryopsis hypnoides) and two mitochondrial genomes (O. viridis and P. akinetum) from the class Ulvophyceae have been published. Here, we present the first chloroplast and mitochondrial genomes from the ecologically and economically important marine, green algal genus Ulva. The chloroplast genome of Ulva sp. was 99,983 bp in a circular-mapping molecule that lacked inverted repeats, and thus far, was the smallest ulvophycean plastid genome. This cpDNA was a highly compact, AT-rich genome that contained a total of 102 identified genes (71 protein-coding genes, 28 tRNA genes, and three ribosomal RNA genes). Additionally, five introns were annotated in four genes: atpA (1), petB (1), psbB (2), and rrl (1). The circular-mapping mitochondrial genome of Ulva sp. was 73,493 bp and follows the expanded pattern also seen in other ulvophyceans and trebouxiophyceans. The Ulva sp. mtDNA contained 29 protein-coding genes, 25 tRNA genes, and two rRNA genes for a total of 56 identifiable genes. Ten introns were annotated in this mtDNA: cox1 (4), atp1 (1), nad3 (1), nad5 (1), and rrs (3). Double-cut-and-join (DCJ) values showed that organellar genomes across Chlorophyta are highly rearranged, in contrast to the highly conserved organellar genomes of the red algae (Rhodophyta). A phylogenomic investigation of 51 plastid protein-coding genes showed that Ulvophyceae is not monophyletic, and also placed Oltmannsiellopsis (Oltmannsiellopsidales) and Tetraselmis (Chlorodendrophyceae) closely to Ulva (Ulvales) and Pseudendoclonium (Ulothrichales).
Conflict of interest statement
Figures
Similar articles
-
De novo assembly of the mitochondrial genome of Ulva fasciata Delile (Ulvophyceae, Chlorophyta), a distromatic blade-forming green macroalga.Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Sep;27(5):3817-9. doi: 10.3109/19401736.2015.1082095. Epub 2015 Sep 15. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26369927
-
The chloroplast genome of the marine green macroalga Ulva fasciata Delile (Ulvophyceae, Chlorophyta).Mitochondrial DNA A DNA Mapp Seq Anal. 2017 Jan;28(1):93-95. doi: 10.3109/19401736.2015.1110814. Epub 2015 Dec 28. Mitochondrial DNA A DNA Mapp Seq Anal. 2017. PMID: 26710262
-
The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes.BMC Biol. 2006 Feb 3;4:3. doi: 10.1186/1741-7007-4-3. BMC Biol. 2006. PMID: 16472375 Free PMC article.
-
Hydrocytium expands the phylogenetic, morphological, and genomic diversity of the poorly known green algal order Chaetopeltidales.Am J Bot. 2023 Nov;110(11):e16238. doi: 10.1002/ajb2.16238. Epub 2023 Oct 24. Am J Bot. 2023. PMID: 37661934 Review.
-
Cytoplasmic inheritance in green algae: patterns, mechanisms and relation to sex type.J Plant Res. 2010 Mar;123(2):171-84. doi: 10.1007/s10265-010-0309-6. Epub 2010 Jan 30. J Plant Res. 2010. PMID: 20112126 Review.
Cited by
-
Chloroplast genomes as a tool to resolve red algal phylogenies: a case study in the Nemaliales.BMC Evol Biol. 2016 Oct 10;16(1):205. doi: 10.1186/s12862-016-0772-3. BMC Evol Biol. 2016. PMID: 27724867 Free PMC article.
-
Screening and verification of extranuclear genetic markers in green tide algae from the Yellow Sea.PLoS One. 2021 Jun 1;16(6):e0250968. doi: 10.1371/journal.pone.0250968. eCollection 2021. PLoS One. 2021. PMID: 34061855 Free PMC article.
-
Large Phylogenomic Data sets Reveal Deep Relationships and Trait Evolution in Chlorophyte Green Algae.Genome Biol Evol. 2021 Jul 6;13(7):evab101. doi: 10.1093/gbe/evab101. Genome Biol Evol. 2021. PMID: 33950183 Free PMC article.
-
Large Diversity of Nonstandard Genes and Dynamic Evolution of Chloroplast Genomes in Siphonous Green Algae (Bryopsidales, Chlorophyta).Genome Biol Evol. 2018 Apr 1;10(4):1048-1061. doi: 10.1093/gbe/evy063. Genome Biol Evol. 2018. PMID: 29635329 Free PMC article.
-
The complete mitochondrial genome sequence of Bryopsis plumosa.Mitochondrial DNA B Resour. 2020 Feb 6;5(1):1067-1068. doi: 10.1080/23802359.2020.1721031. Mitochondrial DNA B Resour. 2020. PMID: 33366877 Free PMC article.
References
-
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, et al. (2012) Phylogeny and Molecular Evolution of the Green Algae. Crit Rev Plant Sci 31: 1–46.
-
- Fučíková K, Leliaert F, Cooper ED, Škaloud P, D'hondt S, De Clerck O, et al. (2014) New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front Ecol Evol 2: 63.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

