Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions
- PMID: 25849964
- PMCID: PMC4388706
- DOI: 10.1371/journal.pone.0122847
Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions
Abstract
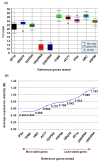
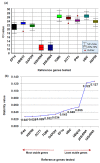
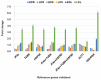
Gene expression analysis using quantitative real-time PCR (qRT-PCR) is a very sensitive technique and its sensitivity depends on the stable performance of reference gene(s) used in the study. A number of housekeeping genes have been used in various expression studies in many crops however, their expression were found to be inconsistent under different stress conditions. As a result, species specific housekeeping genes have been recommended for different expression studies in several crop species. However, such specific housekeeping genes have not been reported in the case of pigeonpea (Cajanus cajan) despite the fact that genome sequence has become available for the crop. To identify the stable housekeeping genes in pigeonpea for expression analysis under drought stress conditions, the relative expression variations of 10 commonly used housekeeping genes (EF1α, UBQ10, GAPDH, 18SrRNA, 25SrRNA, TUB6, ACT1, IF4α, UBC and HSP90) were studied on root, stem and leaves tissues of Asha (ICPL 87119). Three statistical algorithms geNorm, NormFinder and BestKeeper were used to define the stability of candidate genes. geNorm analysis identified IF4α and TUB6 as the most stable housekeeping genes however, NormFinder analysis determined IF4α and HSP90 as the most stable housekeeping genes under drought stress conditions. Subsequently validation of the identified candidate genes was undertaken in qRT-PCR based gene expression analysis of uspA gene which plays an important role for drought stress conditions in pigeonpea. The relative quantification of the uspA gene varied according to the internal controls (stable and least stable genes), thus highlighting the importance of the choice of as well as validation of internal controls in such experiments. The identified stable and validated housekeeping genes will facilitate gene expression studies in pigeonpea especially under drought stress conditions.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

