Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum
- PMID: 25849986
- PMCID: PMC4414887
- DOI: 10.1038/nn.3995
Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum
Abstract
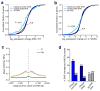
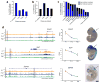
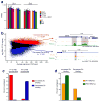
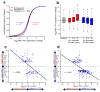
To identify chromatin mechanisms of neuronal differentiation, we characterized chromatin accessibility and gene expression in cerebellar granule neurons (CGNs) of the developing mouse. We used DNase-seq to map accessibility of cis-regulatory elements and RNA-seq to profile transcript abundance across postnatal stages of neuronal differentiation in vivo and in culture. We observed thousands of chromatin accessibility changes as CGNs differentiated, and verified, using H3K27ac ChIP-seq, reporter gene assays and CRISPR-mediated activation, that many of these regions function as neuronal enhancers. Motif discovery in differentially accessible chromatin regions suggested a previously unknown role for the Zic family of transcription factors in CGN maturation. We confirmed the association of Zic with these elements by ChIP-seq and found, using knockdown, that Zic1 and Zic2 are required for coordinating mature neuronal gene expression patterns. Together, our data reveal chromatin dynamics at thousands of gene regulatory elements that facilitate the gene expression patterns necessary for neuronal differentiation and function.
Figures




References
-
- Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

