Interplay between chemotaxis and contact inhibition of locomotion determines exploratory cell migration
- PMID: 25851023
- PMCID: PMC4391292
- DOI: 10.1038/ncomms7619
Interplay between chemotaxis and contact inhibition of locomotion determines exploratory cell migration
Abstract
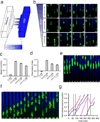
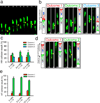
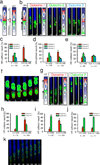
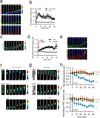
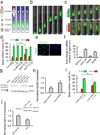
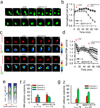
Directed cell migration in native environments is influenced by multiple migratory cues. These cues may include simultaneously occurring attractive soluble growth factor gradients and repulsive effects arising from cell-cell contact, termed contact inhibition of locomotion (CIL). How single cells reconcile potentially conflicting cues remains poorly understood. Here we show that a dynamic crosstalk between epidermal growth factor (EGF)-mediated chemotaxis and CIL guides metastatic breast cancer cell motility, whereby cells become progressively insensitive to CIL in a chemotactic input-dependent manner. This balance is determined via integration of protrusion-enhancing signalling from EGF gradients and protrusion-suppressing signalling induced by CIL, mediated in part through EphB. Our results further suggest that EphB and EGF signalling inputs control protrusion formation by converging onto regulation of phosphatidylinositol 3-kinase (PI3K). We propose that this intricate interplay may enhance the spread of loose cell ensembles in pathophysiological conditions such as cancer, and possibly other physiological settings.
Figures

Similar articles
-
Regulation of contact inhibition of locomotion by Eph-ephrin signalling.J Microsc. 2013 Sep;251(3):232-41. doi: 10.1111/jmi.12024. Epub 2013 Mar 15. J Microsc. 2013. PMID: 23495724 Free PMC article. Review.
-
Epidermal growth factor modulates prostate cancer cell invasiveness regulating urokinase-type plasminogen activator activity. EGF-receptor inhibition may prevent tumor cell dissemination.Thromb Haemost. 2005 May;93(5):964-75. doi: 10.1160/TH04-09-0637. Thromb Haemost. 2005. PMID: 15886816
-
Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells.Nat Cell Biol. 2010 Dec;12(12):1194-204. doi: 10.1038/ncb2122. Epub 2010 Nov 14. Nat Cell Biol. 2010. PMID: 21076414
-
Contact inhibition of locomotion probabilities drive solitary versus collective cell migration.J R Soc Interface. 2013 Sep 18;10(88):20130717. doi: 10.1098/rsif.2013.0717. Print 2013 Nov 6. J R Soc Interface. 2013. PMID: 24047876 Free PMC article.
-
Mechanisms of Neural Crest Migration.Annu Rev Genet. 2018 Nov 23;52:43-63. doi: 10.1146/annurev-genet-120417-031559. Annu Rev Genet. 2018. PMID: 30476447 Review.
Cited by
-
A Thromboxane A2 Receptor-Driven COX-2-Dependent Feedback Loop That Affects Endothelial Homeostasis and Angiogenesis.Arterioscler Thromb Vasc Biol. 2022 Apr;42(4):444-461. doi: 10.1161/ATVBAHA.121.317380. Epub 2022 Mar 3. Arterioscler Thromb Vasc Biol. 2022. PMID: 35236104 Free PMC article.
-
A CDC42-centered signaling unit is a dominant positive regulator of endothelial integrity.Sci Rep. 2017 Aug 31;7(1):10132. doi: 10.1038/s41598-017-10392-0. Sci Rep. 2017. PMID: 28860633 Free PMC article.
-
Bioanalytic Hybrid System Merging 3-Dimensional Cell Culture and Chromatographic Precision for Unprecedented Preclinical Insights in Molecular Imaging.J Nucl Med. 2025 May 1;66(5):813-816. doi: 10.2967/jnumed.124.269133. J Nucl Med. 2025. PMID: 40113219 Free PMC article.
-
Redistribution of Adhesive Forces through Src/FAK Drives Contact Inhibition of Locomotion in Neural Crest.Dev Cell. 2018 Jun 4;45(5):565-579.e3. doi: 10.1016/j.devcel.2018.05.003. Dev Cell. 2018. PMID: 29870718 Free PMC article.
-
Filopodia-based contact stimulation of cell migration drives tissue morphogenesis.Nat Commun. 2021 Feb 4;12(1):791. doi: 10.1038/s41467-020-20362-2. Nat Commun. 2021. PMID: 33542237 Free PMC article.
References
-
- Schmitt AM, et al. Wnt-Ryk signalling mediates medial-lateral retinotectal topographic mapping. Nature. 2006;439:31–37. - PubMed
-
- Condeelis J, Singer RH, Segall JE. THE GREAT ESCAPE: When Cancer Cells Hijack the Genes for chemotaxis and Motility. Annual Review of Cell and Developmental Biology. 2005;21:695–718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

