Deformability in the cleavage site of primary microRNA is not sensed by the double-stranded RNA binding domains in the microprocessor component DGCR8
- PMID: 25851436
- PMCID: PMC4446130
- DOI: 10.1002/prot.24810
Deformability in the cleavage site of primary microRNA is not sensed by the double-stranded RNA binding domains in the microprocessor component DGCR8
Abstract
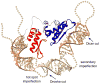
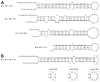
The prevalence of double-stranded RNA (dsRNA) in eukaryotic cells has only recently been appreciated. Of interest here, RNA silencing begins with dsRNA substrates that are bound by the dsRNA-binding domains (dsRBDs) of their processing proteins. Specifically, processing of microRNA (miRNA) in the nucleus minimally requires the enzyme Drosha and its dsRBD-containing cofactor protein, DGCR8. The smallest recombinant construct of DGCR8 that is sufficient for in vitro dsRNA binding, referred to as DGCR8-Core, consists of its two dsRBDs and a C-terminal tail. As dsRBDs rarely recognize the nucleotide sequence of dsRNA, it is reasonable to hypothesize that DGCR8 function is dependent on the recognition of specific structural features in the miRNA precursor. Previously, we demonstrated that noncanonical structural elements that promote RNA flexibility within the stem of miRNA precursors are necessary for efficient in vitro cleavage by reconstituted Microprocessor complexes. Here, we combine gel shift assays with in vitro processing assays to demonstrate that neither the N-terminal dsRBD of DGCR8 in isolation nor the DGCR8-Core construct is sensitive to the presence of noncanonical structural elements within the stem of miRNA precursors, or to single-stranded segments flanking the stem. Extending DGCR8-Core to include an N-terminal heme-binding region does not change our conclusions. Thus, our data suggest that although the DGCR8-Core region is necessary for dsRNA binding and recruitment to the Microprocessor, it is not sufficient to establish the previously observed connection between RNA flexibility and processing efficiency.
Keywords: DGCR8; Drosha; RNA interference; binding affinity; dsRBD; dsRNA; heme binding domain; in vitro processing; microRNA; protein interactions.
© 2015 Wiley Periodicals, Inc.
Figures


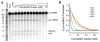




References
-
- Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. - PMC - PubMed
-
- Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nature reviews Drug discovery. 2014;13:622–638. - PubMed
-
- Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review) Molecular medicine reports. 2012;6:9–15. - PubMed
-
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

