The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis
- PMID: 25851604
- PMCID: PMC4472018
- DOI: 10.1091/mbc.E15-02-0073
The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis
Abstract
At the heart of the ribosome lie rRNAs, whose catalytic function in translation is subtly modulated by posttranscriptional modifications. In the small ribosomal subunit of budding yeast, on the 18S rRNA, two adjacent adenosines (A1781/A1782) are N(6)-dimethylated by Dim1 near the decoding site, and one guanosine (G1575) is N(7)-methylated by Bud23-Trm112 at a ridge between the P- and E-site tRNAs. Here we establish human DIMT1L and WBSCR22-TRMT112 as the functional homologues of yeast Dim1 and Bud23-Trm112. We report that these enzymes are required for distinct pre-rRNA processing reactions leading to synthesis of 18S rRNA, and we demonstrate that in human cells, as in budding yeast, ribosome biogenesis requires the presence of the modification enzyme rather than its RNA-modifying catalytic activity. We conclude that a quality control mechanism has been conserved from yeast to human by which binding of a methyltransferase to nascent pre-rRNAs is a prerequisite to processing, so that all cleaved RNAs are committed to faithful modification. We further report that 18S rRNA dimethylation is nuclear in human cells, in contrast to yeast, where it is cytoplasmic. Yeast and human ribosome biogenesis thus have both conserved and distinctive features.
© 2015 Zorbas, Nicolas et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures
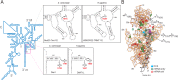
 modifications are conserved in S. cerevisiae and Homo sapiens. (A) Secondary structure of the 18S rRNA. The insets illustrate conservation of rRNA sequence and secondary structure near the N7-methylguanosine (m7G) modification introduced by Bud23-Trm112 in yeast and WBSCR22-TRMT112 in humans and in the vicinity of the two N6,N6-dimethyladenosines
modifications are conserved in S. cerevisiae and Homo sapiens. (A) Secondary structure of the 18S rRNA. The insets illustrate conservation of rRNA sequence and secondary structure near the N7-methylguanosine (m7G) modification introduced by Bud23-Trm112 in yeast and WBSCR22-TRMT112 in humans and in the vicinity of the two N6,N6-dimethyladenosines  synthesized by Dim1 in yeast and DIMT1L in humans. Modified nucleotide positions are highlighted in red. The 5′, central (C), 3′ major (3′ M), and 3′ minor (3′ m) domains are indicated. (B) Three-dimensional representation of the yeast small subunit (model based on Protein Data Bank entry 3U5B) with posttranscriptional modifications and functional sites highlighted. The decoding site (DCS, in cyan) at the base of helix 44 (in anthracite) and the mRNA entry (green circle) and exit (red circle) sites are indicated. Residues shown as green and red spheres are 2′-O methylated or pseudouridylated, respectively. Bd, body; Bk, beak; H, head; Lf, left foot; Nk, neck; Pt; platform, Rf, right foot; Sh, shoulder.
synthesized by Dim1 in yeast and DIMT1L in humans. Modified nucleotide positions are highlighted in red. The 5′, central (C), 3′ major (3′ M), and 3′ minor (3′ m) domains are indicated. (B) Three-dimensional representation of the yeast small subunit (model based on Protein Data Bank entry 3U5B) with posttranscriptional modifications and functional sites highlighted. The decoding site (DCS, in cyan) at the base of helix 44 (in anthracite) and the mRNA entry (green circle) and exit (red circle) sites are indicated. Residues shown as green and red spheres are 2′-O methylated or pseudouridylated, respectively. Bd, body; Bk, beak; H, head; Lf, left foot; Nk, neck; Pt; platform, Rf, right foot; Sh, shoulder.
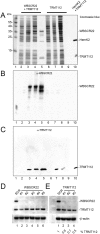



 and m7G1639 methylation, respectively. (A–D) Primer extension mapping of the substrate nucleotides of DIMT1L and WBSCR222 on human 18S rRNA. Total RNA was extracted from HeLa cells depleted for 3 d with a DIMT1L-targeting (A) or WBSCR22-targeting (B) siRNA and analyzed by primer extension with oligonucleotide LD2120, LD2122, or LD2141. (C, D) The positions of the oligonucleotides and of the rRNA species. (E) PNO1 is required for efficient DIMT1L-mediated dimethylation. Total RNA was extracted from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and processed by primer extension with oligonucleotide LD2141. The level of residual dimethylation estimated with a Phosphorimager is indicated to the right. The results shown are means of three independent experiments. The sequences of the siRNAs used are listed in Supplemental Table S3 (WBSCR22#1, DIMT1L#2). (F) PNO1 is not required for the metabolic stability of DIMT1L. Total protein extract from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and tested by Western blotting with the antibodies indicated. Right, residual level of DIMT1L and PNO1 mRNAs assessed by qRT-PCR.
and m7G1639 methylation, respectively. (A–D) Primer extension mapping of the substrate nucleotides of DIMT1L and WBSCR222 on human 18S rRNA. Total RNA was extracted from HeLa cells depleted for 3 d with a DIMT1L-targeting (A) or WBSCR22-targeting (B) siRNA and analyzed by primer extension with oligonucleotide LD2120, LD2122, or LD2141. (C, D) The positions of the oligonucleotides and of the rRNA species. (E) PNO1 is required for efficient DIMT1L-mediated dimethylation. Total RNA was extracted from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and processed by primer extension with oligonucleotide LD2141. The level of residual dimethylation estimated with a Phosphorimager is indicated to the right. The results shown are means of three independent experiments. The sequences of the siRNAs used are listed in Supplemental Table S3 (WBSCR22#1, DIMT1L#2). (F) PNO1 is not required for the metabolic stability of DIMT1L. Total protein extract from HeLa cells depleted for 3 d with a siRNA specific to DIMT1L or PNO1 and tested by Western blotting with the antibodies indicated. Right, residual level of DIMT1L and PNO1 mRNAs assessed by qRT-PCR.

References
-
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. - PubMed
-
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. - PubMed
-
- Brand RC, Klootwijk J, Van Steenbergen TJ, De Kok AJ, Planta RJ. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977;75:311–318. - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

