Krüppel-like Factor 4 Promotes Esophageal Squamous Cell Carcinoma Differentiation by Up-regulating Keratin 13 Expression
- PMID: 25851906
- PMCID: PMC4505602
- DOI: 10.1074/jbc.M114.629717
Krüppel-like Factor 4 Promotes Esophageal Squamous Cell Carcinoma Differentiation by Up-regulating Keratin 13 Expression
Abstract
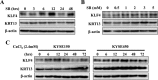
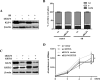
Squamous cell differentiation requires the coordinated activation and repression of genes specific to the differentiation process; disruption of this program accompanies malignant transformation of epithelium. The exploration of genes that control epidermal proliferation and terminal differentiation is vital to better understand esophageal carcinogenesis. KLF4 is a member of the KLF family of transcription factors and is involved in both cellular proliferation and differentiation. This study using immunohistochemistry analysis of KLF4 in clinical specimens of esophageal squamous cell carcinoma (ESCC) demonstrated that decreased KLF4 was substantially associated with poor differentiation. Moreover, we determined that both KLF4 and KRT13 levels were undoubtedly augmented upon sodium butyrate-induced ESCC differentiation and G1 phase arrest. Conversely, silencing of KLF4 and KRT13 abrogated the inhibition of G1-S transition induced by sodium butyrate. Molecular investigation demonstrated that KLF4 transcriptionally regulated KRT13 and the expression of the two molecules appreciably correlated in ESCC tissues and cell lines. Collectively, these results suggest that KLF4 transcriptionally regulates KRT13 and is invovled in ESCC cell differentiation.
Keywords: Kruppel-like factor 4 (KLF4); cell cycle; differentiation; keratin; transcription regulation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Siegel R., Naishadham D., Jemal A. (2012) Cancer statistics, 2012. CA-Cancer J. Clin. 62, 10–29 - PubMed
-
- Ferlay J., Shin H. R., Bray F., Forman D., Mathers C., Parkin D. M. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 - PubMed
-
- Mariette C., Piessen G., Triboulet J. P. (2007) Therapeutic strategies in oesophageal carcinoma: role of surgery and other modalities. Lancet Oncol. 8, 545–553 - PubMed
-
- Segre J. A., Bauer C., Fuchs E. (1999) Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22, 356–360 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

