Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia
- PMID: 25852055
- PMCID: PMC4497970
- DOI: 10.1182/blood-2015-01-620781
Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia
Abstract
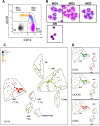
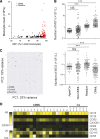
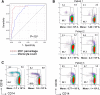
Chronic myelomonocytic leukemia (CMML) is a myelodysplastic syndrome/ myeloproliferative neoplasm whose diagnosis is currently based on the elevation of peripheral blood monocytes to >1 × 10(9)/L, measured for ≥3 months. Diagnosis can be ambiguous; for example, with prefibrotic myelofibrosis or reactive monocytosis. We set up a multiparameter flow cytometry assay to distinguish CD14(+)/CD16(-) classical from CD14(+)/CD16(+) intermediate and CD14(low)/CD16(+) nonclassical monocyte subsets in peripheral blood mononucleated cells and in total blood samples. Compared with healthy donors and patients with reactive monocytosis or another hematologic malignancy, CMML patients demonstrate a characteristic increase in the fraction of CD14(+)/CD16(-) cells (cutoff value, 94.0%). The associated specificity and sensitivity values were 95.1% and 90.6% in the learning cohort (175 samples) and 94.1% and 91.9% in the validation cohort (307 samples), respectively. The accumulation of classical monocytes, which demonstrate a distinct gene expression pattern, is independent of the mutational background. Importantly, this increase disappears in patients who respond to hypomethylating agents. We conclude that an increase in the fraction of classical monocytes to >94.0% of total monocytes is a highly sensitive and specific diagnostic marker that rapidly and accurately distinguishes CMML from confounding diagnoses.
© 2015 by The American Society of Hematology.
Figures


References
-
- Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. - PubMed
-
- Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013;27(7):1441–1450. - PubMed
-
- Ziegler-Heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74–e80. - PubMed
-
- Wong KL, Tai JJ-Y, Wong W-C, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood. 2011;118(5):e16–e31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

