USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus
- PMID: 25852165
- PMCID: PMC4453534
- DOI: 10.1128/mBio.02585-14
USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus
Abstract
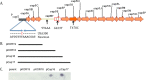
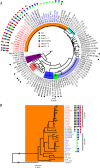
The surface capsular polysaccharide (CP) is a virulence factor that has been used as an antigen in several successful vaccines against bacterial pathogens. A vaccine has not yet been licensed against Staphylococcus aureus, although two multicomponent vaccines that contain CP antigens are in clinical trials. In this study, we evaluated CP production in USA300 methicillin-resistant S. aureus (MRSA) isolates that have become the predominant community-associated MRSA clones in the United States. We found that all 167 USA300 MRSA and 50 USA300 methicillin-susceptible S. aureus (MSSA) isolates were CP negative (CP(-)). Moreover, all 16 USA500 isolates, which have been postulated to be the progenitor lineage of USA300, were also CP(-). Whole-genome sequence analysis of 146 CP(-) USA300 MRSA isolates revealed they all carry a cap5 locus with 4 conserved mutations compared with strain Newman. Genetic complementation experiments revealed that three of these mutations (in the cap5 promoter, cap5D nucleotide 994, and cap5E nucleotide 223) ablated CP production in USA300 and that Cap5E75 Asp, located in the coenzyme-binding domain, is essential for capsule production. All but three USA300 MSSA isolates had the same four cap5 mutations found in USA300 MRSA isolates. Most isolates with a USA500 pulsotype carried three of these four USA300-specific mutations, suggesting the fourth mutation occurred in the USA300 lineage. Phylogenetic analysis of the cap loci of our USA300 isolates as well as publicly available genomes from 41 other sequence types revealed that the USA300-specific cap5 mutations arose sequentially in S. aureus in a common ancestor of USA300 and USA500 isolates.
Importance: The USA300 MRSA clone emerged as a community-associated pathogen in the United States nearly 20 years ago. Since then, it has rapidly disseminated and now causes health care-associated infections. This study shows that the CP-negative (CP(-)) phenotype has persisted among USA300 isolates and is a universal and characteristic trait of this highly successful MRSA lineage. It is important to note that a vaccine consisting solely of CP antigens would not likely demonstrate high efficacy in the U.S. population, where about half of MRSA isolates comprise USA300. Moreover, conversion of a USA300 strain to a CP-positive (CP(+)) phenotype is unlikely in vivo or in vitro since it would require the reversion of 3 mutations. We have also established that USA300 MSSA isolates and USA500 isolates are CP(-) and provide new insight into the evolution of the USA300 and USA500 lineages.
Copyright © 2015 Boyle-Vavra et al.
Figures


References
-
- Trotter CL, McVernon J, Ramsay ME, Whitney CG, Mulholland EK, Goldblatt D, Hombach J, Kieny M-P, SAGE Subgroup . 2008. Optimising the use of conjugate vaccines to prevent disease caused by Haemophilus influenzae type b, Neisseria meningitidis and Streptococcus pneumoniae. Vaccine 26:4434–4445. doi: 10.1016/j.vaccine.2008.05.073. - DOI - PubMed
-
- Hochkeppel HK, Braun DG, Vischer W, Imm A, Sutter S, Staeubli U, Guggenheim R, Kaplan EL, Boutonnier A, Fournier JM. 1987. Serotyping and electron microscopy studies of Staphylococcus aureus clinical isolates with monoclonal antibodies to capsular polysaccharide types 5 and 8. J Clin Microbiol 25:526–530. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

