The neural stem cell fate determinant TRIM32 regulates complex behavioral traits
- PMID: 25852471
- PMCID: PMC4364253
- DOI: 10.3389/fncel.2015.00075
The neural stem cell fate determinant TRIM32 regulates complex behavioral traits
Abstract
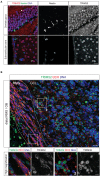
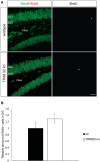
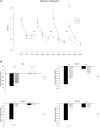
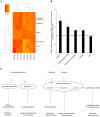
In mammals, new neurons are generated throughout the entire lifespan in two restricted areas of the brain, the dentate gyrus (DG) of the hippocampus and the subventricular zone (SVZ)-olfactory bulb (OB) system. In both regions newborn neurons display unique properties that clearly distinguish them from mature neurons. Enhanced excitability and increased synaptic plasticity enables them to add specific properties to information processing by modulating the existing local circuitry of already established mature neurons. Hippocampal neurogenesis has been suggested to play a role in spatial-navigation learning, spatial memory, and spatial pattern separation. Cumulative evidences implicate that adult-born OB neurons contribute to learning processes and odor memory. We recently demonstrated that the cell fate determinant TRIM32 is upregulated in differentiating neuroblasts of the SVZ-OB system in the adult mouse brain. The absence of TRIM32 leads to increased progenitor cell proliferation and less cell death. Both effects accumulate in an overproduction of adult-generated OB neurons. Here, we present novel data from behavioral studies showing that such an enhancement of OB neurogenesis not necessarily leads to increased olfactory performance but in contrast even results in impaired olfactory capabilities. In addition, we show at the cellular level that TRIM32 protein levels increase during differentiation of neural stem cells (NSCs). At the molecular level, several metabolic intermediates that are connected to glycolysis, glycine, or cysteine metabolism are deregulated in TRIM32 knockout mice brain tissue. These metabolomics pathways are directly or indirectly linked to anxiety or depression like behavior. In summary, our study provides comprehensive data on how the impairment of neurogenesis caused by the loss of the cell fate determinant TRIM32 causes a decrease of olfactory performance as well as a deregulation of metabolomic pathways that are linked to mood disorders.
Keywords: adult neurogenesis; brain metabolism; cell fate determinant; neural stem cells; olfactory behavior.
Figures


References
-
- Belluzzi O., Benedusi M., Ackman J., LoTurco J. J. (2003). Electrophysiological differentiation of new neurons in the olfactory bulb. J. Neurosci. 23, 10411–10418. Available online at: http://www.jneurosci.org/content/23/32/10411.full - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

