Systems modeling approaches for microbial community studies: from metagenomics to inference of the community structure
- PMID: 25852671
- PMCID: PMC4365725
- DOI: 10.3389/fmicb.2015.00213
Systems modeling approaches for microbial community studies: from metagenomics to inference of the community structure
Abstract
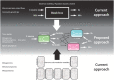
Microbial communities play important roles in health, industrial applications and earth's ecosystems. With current molecular techniques we can characterize these systems in unprecedented detail. However, such methods provide little mechanistic insight into how the genetic properties and the dynamic couplings between individual microorganisms give rise to their dynamic activities. Neither do they give insight into what we call "the community state", that is the fluxes and concentrations of nutrients within the community. This knowledge is a prerequisite for rational control and intervention in microbial communities. Therefore, the inference of the community structure from experimental data is a major current challenge. We will argue that this inference problem requires mathematical models that can integrate heterogeneous experimental data with existing knowledge. We propose that two types of models are needed. Firstly, mathematical models that integrate existing genomic, physiological, and physicochemical information with metagenomics data so as to maximize information content and predictive power. This can be achieved with the use of constraint-based genome-scale stoichiometric modeling of community metabolism which is ideally suited for this purpose. Next, we propose a simpler coarse-grained model, which is tailored to solve the inference problem from the experimental data. This model unambiguously relate to the more detailed genome-scale stoichiometric models which act as heterogeneous data integrators. The simpler inference models are, in our opinion, key to understanding microbial ecosystems, yet until now, have received remarkably little attention. This has led to the situation where the modeling of microbial communities, using only genome-scale models is currently more a computational, theoretical exercise than a method useful to the experimentalist.
Keywords: community modeling; flux balance analysis; genome-scale stoichiometric modeling; metabolism; metagenomic data integration; microbial communities.
Figures



References
-
- Abate A., Hillen R. C., Aljoscha Wahl S. (2012). Piecewise affine approximations of fluxes and enzyme kinetics from in vivo 13C labeling experiments. Int. J. Robust Nonlin. Control 22, 1120–1139 10.1002/rnc.2798 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

