SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data
- PMID: 25852748
- PMCID: PMC4367434
- DOI: 10.3389/fgene.2015.00109
SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data
Abstract
Effective population size (Ne ) is a key population genetic parameter that describes the amount of genetic drift in a population. Estimating Ne has been subject to much research over the last 80 years. Methods to estimate Ne from linkage disequilibrium (LD) were developed ~40 years ago but depend on the availability of large amounts of genetic marker data that only the most recent advances in DNA technology have made available. Here we introduce SNeP, a multithreaded tool to perform the estimate of Ne using LD using the standard PLINK input file format (.ped and.map files) or by using LD values calculated using other software. Through SNeP the user can apply several corrections to take account of sample size, mutation, phasing, and recombination rate. Each variable involved in the computation such as the binning parameters or the chromosomes to include in the analysis can be modified. When applied to published datasets, SNeP produced results closely comparable with those obtained in the original studies. The use of SNeP to estimate Ne trends can improve understanding of population demography in the recent past, provided a sufficient number of SNPs and their physical position in the genome are available. Binaries for the most common operating systems are available at https://sourceforge.net/projects/snepnetrends/.
Keywords: SNPChip; demography; effective population size; large scale genotyping; linkage disequilibrium.
Figures

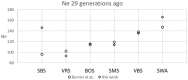
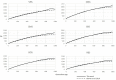
References
-
- Burren A., Signer-Hasler H., Neuditschko M., Tetens J., Kijas J. W., Drögemüller C., et al. (2014). Fine-scale population structure analysis of seven local Swiss sheep breeds using genome-wide SNP data. Anim. Genet. Resour. 55, 67–76 10.1017/S2078633614000253 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

