Statistical approaches to detecting and analyzing tandem repeats in genomic sequences
- PMID: 25853125
- PMCID: PMC4362331
- DOI: 10.3389/fbioe.2015.00031
Statistical approaches to detecting and analyzing tandem repeats in genomic sequences
Abstract
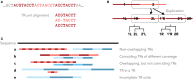
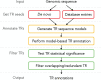
Tandem repeats (TRs) are frequently observed in genomes across all domains of life. Evidence suggests that some TRs are crucial for proteins with fundamental biological functions and can be associated with virulence, resistance, and infectious/neurodegenerative diseases. Genome-scale systematic studies of TRs have the potential to unveil core mechanisms governing TR evolution and TR roles in shaping genomes. However, TR-related studies are often non-trivial due to heterogeneous and sometimes fast evolving TR regions. In this review, we discuss these intricacies and their consequences. We present our recent contributions to computational and statistical approaches for TR significance testing, sequence profile-based TR annotation, TR-aware sequence alignment, phylogenetic analyses of TR unit number and order, and TR benchmarks. Importantly, all these methods explicitly rely on the evolutionary definition of a tandem repeat as a sequence of adjacent repeat units stemming from a common ancestor. The discussed work has a focus on protein TRs, yet is generally applicable to nucleic acid TRs, sharing similar features.
Keywords: molecular evolution; protein domain; sequence profile model; tandem repeat annotation; tandem repeats.
Figures
References
-
- Benson G., Dong L. (1999). Reconstructing the duplication history of a tandem repeat. Proc. Int. Conf. Intell. Syst. Mol. Biol. 44–53. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

